03&04&05 Making Crosses and Establishing the Pedigree Nurseries
From ICISWiki
Contents |
Germplasm Lists: Formation of the Hybridization Block
Introduction:
Once you have selected the parents you wish to cross you need to make a list of those lines for planting in a Hybridization Block. In this exercise, we assume that all the parents are already in the IRIS database. If this is not the case, then these can be entered together with their pedigrees during the formation of the HB or prior to its formation if it is already known which lines are not in the database.
Objectives: At the end of this chapter, the user should be able to:
- Add lines to the HB list.
- Use the IRIS database to form the HB list.
Tutorial Steps:
Forming the HB
The process of forming the HB list is identical to that of forming an OYT or AYT list or any list of existing lines for any purpose. The steps are as follows:
- Select List>New from the List menu. Enter the name for the new list (U03WSHB), enter a title (Upland hybridization block 2003 Wet Season), select the list type (HB) and check the date.
- Click the + button on the tool bar to add lines to the list (see ICIS Technical Documentation\Application Programs\SET GENERATION\1.6 Basic Lists [1]). Enter the first designation (CNA 4196), leave the Entry Code field, but fill the Source (IURON12). Click OK.
- This starts a name search for CNA 4196; if there are several hits, select the one with the IRTP number as the parent since the seed came from INGER IURON nursery.
- Repeat steps 2 and 3 for the other 15 lines in the table below.
| Designation | Cross | GID | Entry Code | Source |
|---|---|---|---|---|
| CNA 4196 | CNA 4196 | 70732 | HB001U | IURON12 |
| IDSA 113 | IDSA 113 | 904702 | HB002U | IURON03 |
| IRAT 170 | IRAT 13/PALAWAN | 677139 | HB003U | IURON15 |
| UPL RI 5 | SIGADIS/BPI 76-1 | 406626 | HB004U | IURON07 |
| WAB 326-B-B-7-H1 | TOX 1785-19-18/WABC 165 | 418229 | HB005U | IURON06 |
| WAB 534-B-3A 1-1 | WAB 181-18/DR 2 | 905029 | HB006U | IURON11 |
| YUNLU NO 28 | IDSA 6/WUNENGDABAIGU-2-5 | 790394 | HB007U | IURON04 |
| APO | UPL RI 5/IR 12979-24-1(BROWN) | 204538 | HB008U | IURON17 |
| IR 72768-12-1-1 | IR 60080-46 A/IR 65907-116-1-B | 1161408 | HB009U | U03DSOYT |
| IR 72768-28-1-1 | IR 60080-46 A/IR 65907-116-1-B | 1161406 | HB010U | U03DSOYT |
| IR 75502-24-1-1-B | B 6144 F-MR-6-0-0/NSIC RC 9 | 1161458 | HB011U | U03DSOYT |
| IR 75516-30-1-1-B | IR 53236-275-1/CT 6516-24-3-2 | 1161444 | HB012U | U03DSOYT |
| IR 75516-56-1-1-B | IR 53236-275-1/CT 6516-24-3-2 | 1161445 | HB013U | U03DSOYT |
| IR 75518-84-1-1-B | IR 60080-46 A/IR 53236-275-1 | 1161448 | HB014U | U03DSOYT |
| IR 75531-31-1-2-B | IR 70360-54-1-B/VIENG | 1161440 | HB015U | U03DSOYT |
| IR 76561-AC 8-B | CT 13382-9-4-M/IR 70358-145-1-1 | 1161327 | HB016U | U03DSOYT |
Now it is possible to fill the Entry Code columns in several ways. We may want to enter a list of Plot Numbers in this field. To do this, right click on the Entry Code column heading and select Fill with Sequence. In the Naming Convention Box enter HB as the Start Constant, 1 in the first plot number, U as the End Constant, check Include Leading Zeros and set Numeric Field Width to 3. Click Ok. The Entry Code field should be filled as in the table above. Save the list, then export it to Excel as HB nursery planting list.
New Germplasm: Specifying Crosses
Introduction:
There are several ways the F1 list can be formed. One may want to specify the particular crosses to be made in advance. At other times, the HB will be checked periodically and crosses could be made according to flowering synchronization. Sometimes users only want to enter their successful crosses. All variations are possible and crosses can be made interactively with the X button on the tool bar (see ICIS Technical Documentation\Application Programs\SET GENERATION\1.9 Cross List[2]). Certain mating designs can in fact be automatically generated with the Matrix Crossing Tool (Mx on the tool bar) and crosses can be constructed from information in a DMS study using the Bx button on the tool bar (see ICIS Tehcnical Documentation\Application Programs\SET GENERATION\1.11 Batch Mode Processing [3]).
Objectives:
At the end of this chapter, the user should be able to:
- Follow the steps to produce a total of 64 planned crosses.
- Use each non-IR line as female parent and each IR line as male parent.
- See that many other combinations are feasible.
Tutorial Steps:
The F1 List
- Start SETGEN from the Launcher (see ICIS Technical Documentation\Application Programs\SET GENERATION\1.2 Starting SetGen[4]).
- Select List>New from the List menu. Enter the name (U04DSF1) and the title (Upland F1 nursery for 2004 Dry Season), select type (F1) and check the date.
- Click the X button on the toolbar. Double click U03WSHB in the panel of existing lists. This will open the HB in the browse window of the crossing tool.
- Select Cross Name as the Name Type and check the date of crossing. Specify the crossing location by clicking <Change next to the Name Location box. Select Philippines from the Country box, and type IRRI-I to move the location selector to IRRI-International Rice Research Institute. Double click this entry to select it.
- Check the Use Naming Convention box. Enter IR in the Start Constant box, and 90599 in the Next Integer box. Click Ok.
- Click CNA 4196 in the browse window and then click Female to specify this as the Female parent.
- Select all the IR lines in the browse window by clicking each in turn or dragging the mouse over them (The last eight entries in the HB list). Click the male button to specify the first of these as the male parent.
- Click Next to proceed to the next selected Male parent repeat for all the remaining male parents.
- Repeat steps 5 to 7 for each female line (non-IR lines) to generate 64 crosses in all.
Now right click on the Entry Code column heading and select Fill with Sequence. In the Naming Convention box enter P as the start constant, 1 as the first plot number, leave the end constant blank but check Include Leading Zeros and set numeric field width to 3. Click Ok. The Entry Code field should be filled with the sequence P001 to P064. Save the list, then export it to Excel as an F1 nursery planting list.
The F2 Nursery
We assume that F2 seeds from all F1 plants in each F1 plot are bulked and then space planted in plots for the F2 nursery. This means that the F2 Nursery planting list can look exactly like the F1 planting list.
To make the F2 nursery list, simply copy the F1 list and enter new plot identifiers as follows:
- Open the F1 list (select U04DSF1) in the Browse Window (right click).
- Select List>New from the List menu. Enter the name (U04WSF2) and the title (Upland F2 nursery for 2004 Wet Season), type (F2) and verify that the date is correct.
- Tag all the lines in the F1 list in the Browse window (Right click on the Tag column heading).
- Click the Derive lines button (-), select Random Bulk as the method, select Derivative Name as the name type, leave the Start constant field of the Naming Convention section, complete the date and locations section, and click OK to All.
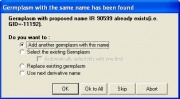
- A small confirmation box will appeared saying “No naming convention specified. Is the derivative name same as its source?”. Click Yes.
- An information window displays notifying the user that a germplasm with the same name was found.
- Select ‘Add another germplasm with this name’ to add the same germplasm name but generate a different GID to the list.
- Click on ‘Ok to All’. This will generate new GIDs with the same germplasm names for all of the items in the list.
- In the new list (U04WSF2), right click on the Entry Code column heading and select Fill with Sequence. In the Naming Convention Box enter B as the start constant, 1 in Next Integer box and the first plot number, enter F2 as the end constant, check Include Leading Zeros and set numeric field width to 3. Click Close. The Entry Code field should be filled with the sequence B001F2 to B064F2. Save the list, then export it to Excel as an F2 nursery planting list.
Table of ICIS Breeding Methods for Self Fertilizing Crops
Methods for storing historical pedigrees with incomplete information
| METHN | MTYPE | MGRP | MCODE | MNAME | MDESC |
|---|---|---|---|---|---|
| 1 | GEN | S | UGM | UNKNOWN GENERATIVE METHOD SF | Unknown generative method for storing historic pedigrees for self fertilizing species. |
| 4 | GEN | S | BDU | F1 BACKCROSS, CYTOPLASM UNKNOWN SF | Cross of F1 to recurrent parent when the direction of the cross is unknown for storing historic pedigrees for self fertilizing species. |
| 6 | GEN | S | BRU | F2 BACKCROSS, CYTOPLASM UNKNOWN SF | Cross of F2 to recurrent parent when the direction of the cross is unknown for storing historic pedigrees for self fertilizing species. |
| 8 | GEN | G | CCU | CROSS, CYTOPLASM UNKNOWN | Cross between two plants, unknown which is female |
| 31 | DER | S | UDM | UNKNOWN DERIVATIVE METHOD SF | Unknown derivative method in self-fertilizing species: for storing historic pedigrees |
Generic Maintenance Methods
| METHN | MTYPE | MGRP | MCODE | MNAME | MDESC |
|---|---|---|---|---|---|
| 60 | MAN | G | IDN | PLANT IDENTIFICATION | Identifying and naming a plant or population. |
| 61 | MAN | G | NSI | SEED INCREASE | Increase seed of a cultivar, line, population or accession. |
| 62 | MAN | G | ISE | IMPORT | Import seed, clones or tissue culture of a cultivar, line, population or accession. |
| 63 | MAN | G | ESE | EXPORT | Export seed, clones or tissue culture of a cultivar, line, population or accession. This method is not required. |
| 64 | MAN | G | SSN | STORE SEED NORMAL | Store seed of a cultivar, line, population or accession in normal method: drift not expected. It is unlikely that this method is needed. |
| 65 | MAN | G | SSM | STORE SEED MEDIUM TERM | Store seed of a cultivar, line, population or accession in medium term storage. Some genetic drift is expected. Storage is between 0-4OC and low RH. |
| 66 | MAN | G | SSL | STORE SEED LONG TERM | Store seed of a cultivar, line, population or accession. Genetic drift is expected. Storage is about -18OC. |
Generative Methods for Inbreeding Crops
| METHN | MTYPE | MGRP | MCODE | MNAME | MDESC |
|---|---|---|---|---|---|
| 101 | GEN | S | C2W | SINGLE CROSS (may consider adding TEST CROSS, for hybrid rice breeding) | Cross between two single plants. If both parents are fixed (pure) inbred lines there will be no segregation for gametes or genotypes and theoretically all crosses will result in the same genetic outcome. In plant breeding practice the theoretical situation is rarely encountered. In spite of this the usual practice is to bulk the seed. However, in genetical studies it is often necessary to keep individual seed separate. When this is done a separate entry in the germplasm table is required for each entity (seed) kept separate. |
| 102 | GEN | S | C3W | THREE-WAY CROSS | Cross between two plants, one an inbred line and one a single cross (usually an F1) and thus segregating for gametes. In the theoretical case, rarely achieved, the inbred line would be fixed and the F1 a cross between fixed lines. The segregation for gametes results in different genetic outcomes among different progeny, hence a number of crosses using the same F1 is usually made. Since different F1 s are genetically the same (theoretically) only one F1 is required. In plant breeding programs the different crosses are usually bulked. Again, if individual seeds are kept separate a different entry is required in the germplasm table. |
| 103 | GEN | S | CDB | DOUBLE CROSS | Cross between two single crosses (usually two F1s) and hence both segregating for gametes. The comments for method 102 apply but now for both female and male sides of the cross. Again, if individual seeds are kept separate a different entry is required in the germplasm table. |
| 104 | GEN | S | CFT | FEMALE COMPLEX TOP CROSS | Cross between a female inbred line and a three-way or more complex cross among inbred lines, thus the male is segregating for genotypes as well as gametes. A consequence of the genotypic segregation is that selection can, and is usually made among the plants used as male parents. A consequence is that there will be genetic variation both within and between each cross. Usually all seed is bulked and selection practiced among the progeny. A different entry is required in the germplasm table for each entity kept separate. |
| 105 | GEN | S | CMT | MALE COMPLEX TOP CROSS | Cross between a male inbred line and a three-way or more complex cross among inbred lines, thus the female is segregating for genotypes as well as gametes. The same genetic consequences occur as for the previous complex cross except for the cytoplasm. This method is rarely if ever encountered in practice because of the difficulty of using many females. A different entry is required in the germplasm table for each entity kept separate. |
| 106 | GEN | S | CCX | COMPLEX CROSS | Cross between two three-way or more complex crosses among pure lines, thus both sides are segregating for both gametes and genotypes. A different entry is required in the germplasm table for each entity kept separate. |
| 107 | GEN | S | BC | BACKCROSS | Backcross to recover a specific gene. The coding in the genealogical table records which parent was used as the female in each cycle. A different entry is required in the germplasm table for each entity kept separate. |
| 108 | GEN | S | BCR | BACKCROSS RECESSIVE | Backcross to recover a recessive gene. As this requires a self fertilization (derivative method) in the process some ICIS administrators may distinguish this as a separate method. A different entry is required in the germplasm table for each entity kept separate. |
| 109 | GEN | S | CIS | INTERSPECIFIC CROSS | Cross between two species. The problem with making this a separate method is that the species cross could be made by any of the previous (101-108) or following (110-113) methods. |
| 110 | GEN | S | CSP | SELECTED POLLEN CROSS SF | A bulk of pollen from a selected set of males used to pollinate a female inbred line. |
| 111 | GEN | S | CRP | RANDOM POLLEN CROSS SF | A random bulk of pollen from some population used to pollinate a female pure line. Male is then a population and will be recorded as a single entity. |
| 112 | GEN | S | CGO | OPEN POLLENATED SF | Open pollination in a self- fertilized species |
| 151 | GEN | S | MUN | NATURAL VARIANT SF | A recognized naturally occurring variant in a self- fertilizing population. |
| 152 | GEN | S | MIP | INDUCED MUTATION POPULATION SF | A population derived from inducing mutation in a inbred line. |
| 153 | GEN | S | SCL | SOMACLONE SF | Variation induced through tissue culture of a inbred line. |
| 154 | GEN | S | ALP | ALLOPOLYPLOID SF | Polyploid formed by doubling the chromosomes of a cross between two or more species. Wheat is an allopolyploid as it contains genomes from three different species. |
| 155 | GEN | S | AUP | AUTOPOLYPLOID SF | Polyploid formed by doubling the chromosome number of a species. Lucerne (alfalfa) is an autopolyploid with 4 sets of the same genome. |
| 156 | GEN | S | HAP | HAPLOID SF | Individual with chromosome content of reduced gamete. Often formed by female progenitors crossed with a haploid inducer. |
| 157 | GEN | S | TRN | TRANSGENIC NUCLEUS SF | Individual derived from genetic transformation of the nucleus in a self fertilizing species. |
| 158 | GEN | S | TRC | TRANSGENIC CYTOPLASM SF | Individual derived from genetic transformation of a cytoplasm inclusion (e.g. chloroplast) in a self- fertilizing species. |
Derivative Methods for Inbreeding Crops
| METHN | MTYPE | MGRP | MCODE | MNAME | MDESC |
|---|---|---|---|---|---|
| 201 | DER | S | MIL | INDUCED MUTATION LINE | A recognized mutation selected from an induced mutation in a line of a self-fertilized species. |
| 202 | DER | S | DDH | DOUBLE HAPLOID LINE | Individual produced by doubling haploid individual usually by anther culture in a self- fertilized crop. |
| 203 | DER | S | DPR | PURIFICATION | Selection of one or a few plants from an inbred line or pure line cultivar. |
| 204 | DER | S | DRU | ROUGING SF | Eliminating off types from a inbred line or pure line cultivar. |
| 205 | DER | S | DSP | SINGLE PLANT SELECTION SF | Derivation through selection of a single plant, inflorescence, fruit or seed from a self-fertilizing population. |
| 206 | DER | S | DSB | SELECTED BULK SF | Derivation through bulking seed from a selected set of single plants from a self-fertilizing population. |
| 207 | DER | S | DRB | RANDOM BULK SF | Derivation through bulking seed from a random selection of single plants from a self-fertilizing population. |
| 208 | DER | S | DSD | SINGLE SEED DESCENT SF | Derived through the production of a single individual without selection from each individual in a segregating population. |
| 209 | DER | S | DRS | CMS RESTORER SELECTION | Restorer Lines selected at the end of a program to back cross a gene which restores male fertility to lines carrying a Male Sterile Cytoplasm (CMS) to the male of a commercial hybrid. |
| 210 | DER | S | DMS | CMS MAINTAIN ER SELECTION | Maintainer line selected at the end of a program to create the male fertile equivalent of the CMS female parent of a hybrid |
| 251 | DER | S | ALP | LANDRACE POPULATION SF | Acquisition only.
A Landrace Accession of a self-fertilized species. This population will consist of a heterogenous mixture of homogenous genotypes. This and the following eight methods should be reserved for the acquisition of these types of population to any program when they are first collected. When they are transferred from one collection (germplasm bank, working collection or plant improvement program) to another they should be entered under the IMPORT method. |
| 252 | DER | S | ALL | LANDRACE LINE SF | Acquisition only.
When the accession derives from a single plant in the Landrace Population. |
| 253 | DER | S | ALC | LANDRACE CULTIVAR SF | Acquisition only.
A Landrace Cultivar Accession of a self-fertilized species. Accession of a long term cultivar, not bred or maintained by modern breeding methods. This would usually be less heterogenous than a traditional landrace. |
| 254 | DER | S | ACP | COLLECTION POPULATION SF | Acquisition only.
An accession of a population of a cultivated self -fertilizing species not from farmer’s fields. |
| 255 | DER | S | ACL | COLLECTION LINE SF | Acquisition only.
When the accession derives from a single plant in a Collection Population. |
| 256 | DER | S | AWP | COLLECTION WILD SPP POPULATION SF | Acquisition only.
An accession of a self-fertilizing species. |
| 257 | DER | S | AWL | COLLECTION WILD SPP LINE SF | Acquisition only.
When the accession derives from a single plant from a wild collection. |
| 258 | DER | S | ADP | COLLECTION WEEDY SPP POPULATION SF | Acquisition only.
An accession of a self-fertilizing species which is a weed (because of the result of a hybrid between the cultivated and a wild species of the crop). |
| 259 | DER | S | ADL | COLLECTION WEEDY SPP LINE SF | Acquisition only.
When the accession derives from a single plant in a collection of weedy species. |
Management Methods for Inbreeding Crops
| METHN | MTYPE | MGRP | MCODE | MNAME | MDESC |
|---|---|---|---|---|---|
| 301 | MAN | S | NSP | SEED INCREASE PLANT SF | Seed increase from a single plant in a self-fertilized species. |
| 302 | MAN | S | NMX | SEED INCREASE MIXTURE SF | Seed increase from a number of selected plants in a self- fertilized species. |
| 303 | MAN | S | NBK | SEED INCREASE BULK SF | Seed increase from an unselected bulk in a self-fertilizing species. |
| 320 | MAN | S | VPL | PURE LINE FORMATION | Forming a pure line CV in a self-fertilizing species. |
| 321 | MAN | S | VHY | HYBRID FORMATION SF | Forming a hybrid CV in a self-fertilizing crop. |
| 322 | MAN | S | VML | MULTI-LINE FORMATION SF | Forming a multi-line CV in a self-fertilizing crop |
| 323 | MAN | S | VBS | BREEDERS SEED SF | Producing Breeder’s Seed. Pure seed produced by breeder (usually some kept by breeder) in a self-fertilizing crop. |
| 324 | MAN | S | VFS | FOUNDATION SEED SF | Producing Foundation Seed. Pure seed derived from Breeders seed (usually kept by seed producing organization) in a self-fertilizing crop. |
| 325 | MAN | S | VCS | CERTIFIED SEED | Producing Certified Seed. Pure seed produced under supervision by Government Protocols. |
| 326 | MAN | S | VCR | CULTIVAR RELEASE | Release a cultivar |