Data Validation Tool 5.5.1
From ICISWiki
M.C. Habito, A.R. Llorca, R. Sackville Hamilton
ICIS Technical Documentation 5.5 > Data Validation Tool 5.5 > Data Validation Tool 5.5.1
Download
Latest version is here: Data Validate 5.5.1.1
Introduction
The ICIS Data Validation Tool is an application that searches ICIS for data errors that might render it meaningless. This is useful in making sure that published data are always of excellent quality.
Just choose the tests you want to execute and click on the "Run" button. If you wish to run several or all tests, it is recommended to leave this tool running overnight.
Genealogy Management System
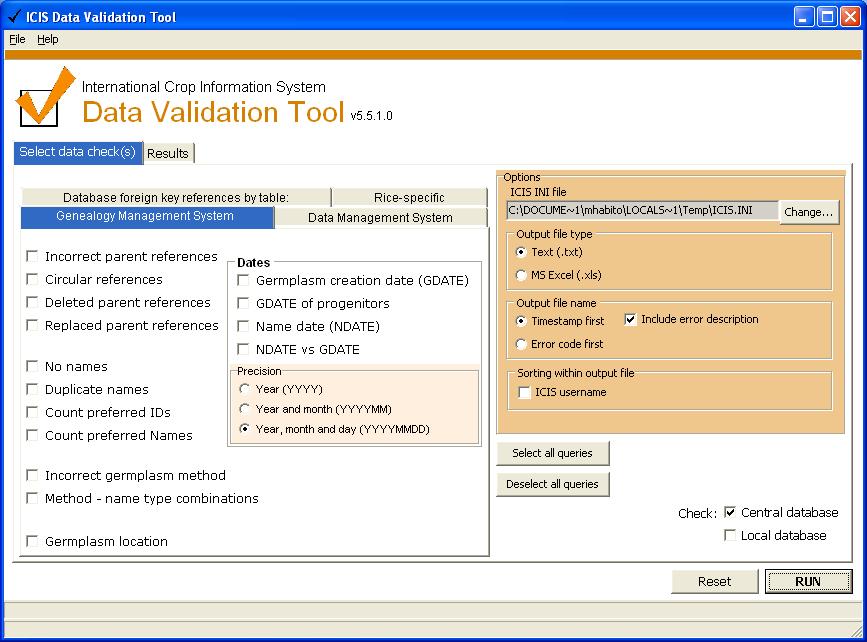
Checks the GMS database (applicable to all ICIS implementations) --> see screenshot above.
Incorrect parent references [2 checks]
| Summary | Error Message | Notes |
|---|---|---|
| Query GIDs in the GMS.GERMPLSM table that have unknown group source, and with non-generative group source. | DataError-0001: Unknown group source | CHK01E |
| DataError-0002: Germplasm with non-generative group source |
Circular references [3 checks]
| Summary | Error Message | Notes |
|---|---|---|
| If germplasm A has germplasm B as one of its parent and if germplasm B has germplasm A as one of its parents, then we have a circular reference situation. This option also checks for two and three-level circularity. | DataError-0003: 1st level circular reference | A references B, and B references A [CHK01C,CHK01D] |
| DataError-0004: 2nd level circular reference | A references B, and B references C, and C references A | |
| DataError-0005: 3rd level circular reference | A references B, and B references C, and C references D, and D references A |
Deleted parent references [3 checks]
| Summary | Error Message | Notes |
|---|---|---|
| A GID should not reference a deleted GPID1, GPID2 or MGID. | DataError-0007A: Germplasm with deleted parent references (GPID1) | |
| DataError-0007B: Germplasm with deleted parent references (GPID2) | ||
| DataError-0007C: Germplasm with deleted parent references (MGID) |
Replaced parent references [3 checks] 
| Summary | Error Message | Notes |
|---|---|---|
| Let us say germplasm A is replaced with germplasm B as shown in the GERMPLSM.GRPLCE database column. All germplasm that references germplasm A should be corrected to germplasm B. | DataError-0015A: Germplasm with replaced parent references (GPID1) | |
| DataError-0015B: Germplasm with replaced parent references (GPID2) | ||
| DataError-0015C: Germplasm with replaced parent references (MGID) |
No names [1 check] 
Checks germplasm with no NAMES records.
Duplicate Names [2 checks] 
CropForge Feature Request # 710 (rhamilton,mhabito)
| Summary | Error Message | Notes |
|---|---|---|
| NAMES table: GID-NVAL-NTYPE (Germplasm ID - Name value - Name type) should be unique in any implementation of ICIS. | DataError-0032: Germplasm with duplicate names | |
| Possible duplicates. NVALs may only have slight differences in spelling | DataError-0010: Germplasm with possible duplicate names (NTYPE-NSTAT combination occurring more than once) |
Count preferred IDs [2 checks]
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| No more (or less) than one name of a GID can have a "preferred ID" status (NSTAT =8) | DataError-0011: Germplasm with no preferred ID (NSTAT=8) | |
| DataError-0020: Germplasm with multiple preferred IDs (NSTAT=8) |
Count preferred names [2 checks]
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| No more (or less) than one name of a GID can have a "preferred name" status (NSTAT =1) | DataError-0017: Germplasm with multiple preferred English names (NSTAT=1) | |
| DataError-0018: Germplasm with no preferred English name (NSTAT=1) |
Incorrect germplasm method [2 checks]
| Summary | Error Message | Notes |
|---|---|---|
| The method of genesis for the germplasm (METHN) should have a corresponding value for number of progenitors (GNPGS) | DataError-0006A: Incorrect germplasm method (Method = DER but GNPGS <> -1) | For a derivative process (Method = DER), GNPGS must be -1 [CHK01A] |
| DataError-0006B: Incorrect germplasm method (Method = GEN but GNPGS < 0) | For a generative process (Method = GEN), GNPGS must be greater than 0 [CHK01B] |
Method - name type combinations [3 checks]
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
Only certain combinations of name type and germplasm creation method are acceptable. Namely:
| Summary | Error Message | Notes |
|---|---|---|
| Method type GEN: valid name types are: CRSNM, UNCRS, UNRES | DataError-0024: Invalid method-name type combination for method GEN (generative) | |
| Method type DER: valid name types are: RELNM, DRVNM, CVNAM, CVABR, NTEST, LNAME, ADVNM, ACVNM, AABBR, OLDMUT1, OLDMUT2, ELITE, UNRES | DataError-0025: Invalid method-name type combination for method DER (derivative) | |
| Method type MAN: valid name types are: ACCNO, RELNM, CVNAM, CVABR, COLNO, FACCN, ITEST, NTEST, LNAME, TACC, ADVNM, ACVNM, ELITE, GACC, DACCN, LCNAM, CIATGB | DataError-0026: Invalid method-name type combination for method MAN (maintenance) |
Germplasm location
| Summary | Error Message | Notes |
|---|---|---|
| For all germplasm with germplasm creation NOT method ID 62 (Import), germplasm location (GLOCN) of a GID should be the same as the GLOCN of its GPID2
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki | DataError-0023B: Germplasm with GLOCN different from GLOCN of GPID2 (METHOD <> IMPORT) |
DATES
Germplasm creation date (GDATE) [3 checks] 
| Summary | Error Message | Notes |
|---|---|---|
| Check for existence/format/value of germplasm creation date (GDATE) | DataError-0034A: Germplasm with METHOD = IMPORT but with no germplasm creation date | Imported germplasm require a germplasm creation date |
| DataError-0034B: Germplasm with incorrect creation date (GDATE) format | If GDATE is not 0, there should be exactly 8 digits and the format should be YYYYMMDD | |
| DataError-0034C: Germplasm with creation date (GDATE) after current date |
GDATE of progenitors [2 checks]
CropForge Feature Request # 160 (rhamilton) and/or Check #1: Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| The GDate of a GID must not predate GDate of any of its progenitors. (Beware of missing links in the chain of dates! If the test checks only the GDATE of the progenitors GPID1, GPID2 and MGID, then it will not detect the following error: non-zero GPID1, GPID2 or MGID has GDATE=0 but their GPID1, GPID2 or MGID are younger than the target GID. Therefore, if a GPID1, GPID2 or MGID has GDate=0, iterate to check their GPID1, GPID2 and MGID) | DataError-0012: Germplasm with germplasm creation date (GDATE) earlier than GDATE of parent (GPID1, GPID2 or both) | |
| DataError-0013: Germplasm with germplasm creation date (GDATE) earlier than GDATE of MGID |
Name date [2 checks]
- DataError-0035A: Germplasm with incorrect name date (NDATE) format
- DataError-0035B: Germplasm with NAME date (NDATE) after current date
NDATE vs GDATE
CropForge Feature Request #631 (jnorgaard)
Name Date (NAMES.NDATE) could also be checked with the Germplasm Date (GERMPLSM.GDATE) to make sure it is not older. SQL modified to check only non-inherited names.
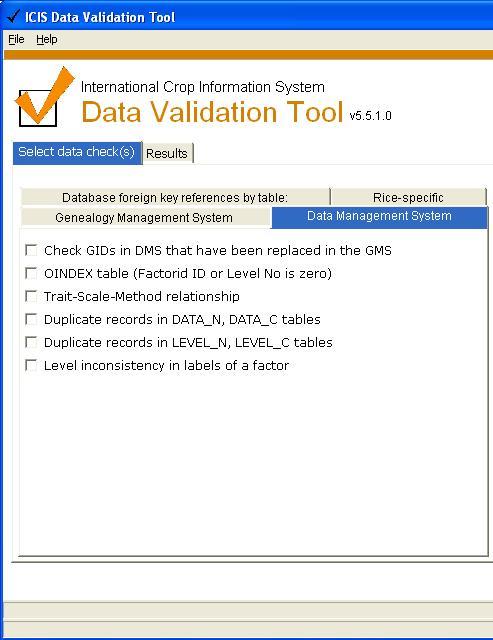
Data Management System
Checks the DMS database (applicable to all ICIS implementations).
Check GIDs in DMS that have been replaced in the GMS 
CropForge Feature Request # 935 (aportugal)
OINDEX table (Factor ID or Level No is zero) 
(Requested by: tmetz, aportugal)
Trait-Scale-Method relationship [4 checks]
CropForge Feature Request # 610 (aportugal)
Check the consistency of the trait, scale and method in the FACTOR and VARIATE tables. The Scale and Method specified in those tables should be related to the the Trait. This can be determine when the traitid in the factor/variate is the same as the traitid in the SCALE and TMETHOD tables for the entered scaleid/tmethid.
DataError-0031A: SCALE.TRAITID is not equal to FACTOR.TRAITID (given the same SCALEID) DataError-0031B: TMETHOD.TRAITID is not equal to FACTOR.TRAITID (given the same TMETHID) DataError-0031C: SCALE.TRAITID is not equal to VARIATE.TRAITID (given the same SCALEID) DataError-0031D: TMETHOD.TRAITID is not equal to VARIATE.TRAITID (given the same TMETHID)
Duplicate records in LEVEL_N, LEVEL_C tables 
(Requested by: tmetz, aportugal)
Duplicate records in DATA_N, DATA_C tables 
(Requested by: tmetz, aportugal)
Level inconsistency in labels of a factor 
(Requested by: tmetz, aportugal)
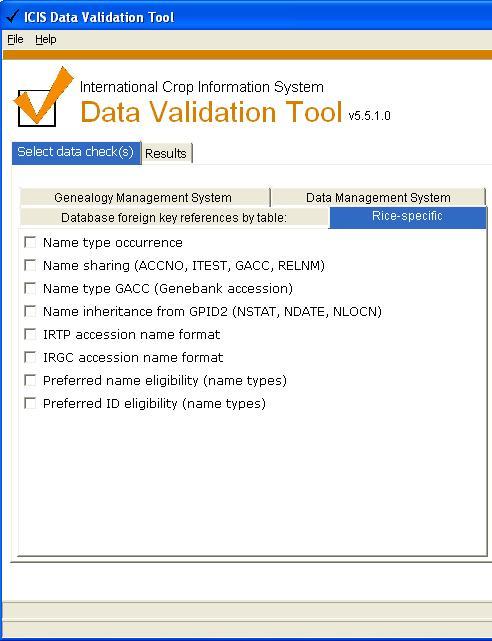
Rice-specific
Checks for Rice.
Name type occurrence [3 checks]
CropForge Feature Request # 160 (rmahilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| Some name types must not occur more than once for a single GID. These are ACCNO, CRSNM, UNCRS, COLNO, ITEST, GACC, CIATGB, RELNM, DRVNM, CVNAM | DataError-0027: Germplasm with certain name types occurring more than once | |
| Cross-names (CRSNM) and Line-names (LNAME) cannot occur together | DataError-0028: Germplasm with both CRSNM and LNAME name types | |
| Release names (RELNM) and Collector's Numbers (COLNO) cannot occur together | DataError-0029: Germplasm with both RELNM and COLNO name types |
Name sharing [4 checks]
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| Germplasms cannot share the same name (NVAL) if the NTYPE is ACCNO | DataError-0030A: Multiple germplasms sharing the same name (ACCNO) | |
| Germplasms cannot share the same name (NVAL) if the NTYPE is ITEST | DataError-0030B: Multiple germplasms sharing the same name (ITEST) | |
| Germplasms cannot share the same name (NVAL) if the NTYPE is GACC | DataError-0030C: Multiple germplasms sharing the same name (GACC) | |
| A Release Name (RELNM) cannot occur more then once for a GID-GLOCN-COUNTRY combination. Two GIDs can share the same RELNM only if their GLOCNs are in different countries | DataError-0030D: Germplasm sharing a RELNM (release name) with another germplasm in the SAME country |
Name type GACC (Genebank accession) / IRGC accession [5 checks] 
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
All names of type GACC must have...
| Summary | Error Message | Notes |
|---|---|---|
| ...a METHN with MTYPE = MAN | DataError-0036A: Genebank accessions (name type GACC) with MTYPE <> Management (MAN) | |
| ...MGID <> 0 | DataError-0036B: Genebank accessions (name type GACC) with MGID = 0 | |
| ...GPID1=MGID if its MGID has GPID1=0 | DataError-0036C: Genebank accessions (name type GACC) with GPID1 <> MGID.
(If a germplasm has an MGID with GPID1=0, then a GID's GPID1 and MGID must be equal) | |
| ...GPID1=[GPID1 of MGID] if [GPID1 of MGID] is not 0 | DataError-0036D: Genebank accessions (name type GACC) with GPID1 <> MGID'S GPID1
(If a germplasm has an MGID with GPID1 not 0, then GPID1 and the MGID's GPID1 must be equal) | |
| IRGC accession must not have a cross name | DataError-0036E: International Rice Genebank Collection (IRGC) accession with Cross Name (NTYPE = 2) |
Name inheritance from GPID2 (NSTAT, NDATE, NLOCN) [3 checks]
CropForge Feature Request # 160 (rhamilton) and/or Check #2: Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| If a GID inherits a name from its source (GPID2) then that name record must also inherit the name date (NDATE), name location (NLOCN), and preferred name status (NSTAT=1) | DataError-0014: MAN germplasm with inherited name from GPID2 but NSTAT not inherited | |
| DataError-0015: Germplasm with inherited name from GPID2 but NDATE not inherited | ||
| DataError-0016: Germplasm with inherited name from GPID2 but NLOCN not inherited |
IRTP name format 
(Requested by: aportugal)
If NSTAT=1, check if everything that follows "IRTP" is numeric.
IRGC name format 
(Requested by: aportugal)
If NSTAT=1, check if everything that follows "IRGC" is numeric.
Preferred name eligibility (name types)
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| Names eligible to be the preferred name are: CRSNM, RELNM, DRVNM, CVNAM, ELITE | DataError-0019: Germplasm with invalid name type as preferred name | Only checks germplasm with more than one NAMES record. |
Preferred ID eligibility (name types) [2 checks]
CropForge Feature Request # 160 (rhamilton) and/or Version 5.3 Discussion Article on ICISWiki
| Summary | Error Message | Notes |
|---|---|---|
| The following names types are eligible to be preferred ID: DRVNM, COLNO, ACCNO (if present), GACC (if present), ITEST (if present), CIATGB (if present) | DataError-0021: Germplasm with invalid name type as preferred ID | Only checks germplasm with more than one NAMES record |
| The preferred ID must be unique for the given name type – that is, two accessions must not share the same name of the same type if one or both is a preferred ID. | DataError-0022: Germplasm with preferred ID not unique for the given name type |
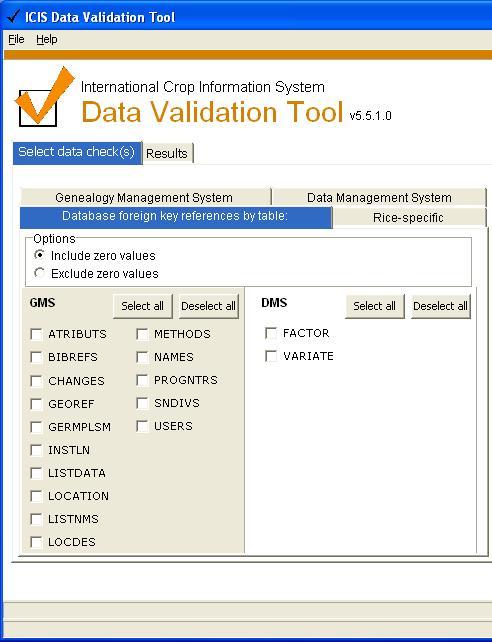
Database foreign key references by tables 
The ICIS database has no foreign key constraints enabled so we need to check if the foreign key definition is violated. Check for foreign key references used by tables:
Miscellaneous Features
- "About" form containing more information, plus the GNU General Public License. CRIL and IRRI logos also included.
- Application icon
DataError Index Listing
- DataError-0001: Unknown group source
- DataError-0002: Germplasm with non-generative group source
- DataError-0003: 1st level circular reference
- DataError-0004: 2nd level circular reference
- DataError-0005: 3rd level circular reference
- DataError-0006A: Incorrect germplasm method (Method = DER but GNPGS <> -1)
- DataError-0006B: Incorrect germplasm method (Method = GEN but GNPGS < 0)
- DataError-0007A: Germplasm with deleted parent references (GPID1)
- DataError-0007B: Germplasm with deleted parent references (GPID2)
- DataError-0007C: Germplasm with deleted parent references (MGID)
- DataError-0008: Germplasm with no NAMES records
- DataError-0009: Germplasm with duplicate names (GID-NVAL-NTYPE not unique)
- DataError-0010: Germplasm with possible duplicate names (NTYPE-NSTAT combination occurring more than once)
- DataError-0011: Germplasm with no preferred ID (NSTAT=8)
- DataError-0012: Germplasm with germplasm creation date (GDATE) earlier than GDATE of parent (GPID1, GPID2 or both)
- DataError-0013: Germplasm with germplasm creation date (GDATE) earlier than GDATE of MGID
- DataError-0014: MAN germplasm with inherited name from GPID2 but NSTAT not inherited
- DataError-0015: Germplasm with inherited name from GPID2 but NDATE not inherited
- DataError-0016: Germplasm with inherited name from GPID2 but NLOCN not inherited
- DataError-0017: Germplasm with multiple preferred English names (NSTAT=1)
- DataError-0018: Germplasm with no preferred English name (NSTAT=1)
- DataError-0019: Germplasm with invalid name type as preferred name
- DataError-0020: Germplasm with multiple preferred IDs (NSTAT=8)
- DataError-0021: Germplasm with invalid name type as preferred ID
- DataError-0022: Germplasm with preferred ID not unique for the given name type
- DataError-0023: Germplasm with GLOCN different from GLOCN of GPID2 (METHOD <> IMPORT)
- DataError-0024: Invalid method-name type combination for method GEN (generative)
- DataError-0025: Invalid method-name type combination for method DER (derivative)
- DataError-0026: Invalid method-name type combination for method MAN (maintenance)
- DataError-0027: Germplasm with certain name types occurring more than once
- DataError-0028: Germplasm with both CRSNM and LNAME name types
- DataError-0029: Germplasm with both RELNM and COLNO name types
- DataError-0030A: Multiple germplasms sharing the same name (ACCNO)
- DataError-0030B: Multiple germplasms sharing the same name (ITEST)
- DataError-0030C: Multiple germplasms sharing the same name (GACC)
- DataError-0030D: Germplasm sharing a RELNM (release name) with another germplasm in the SAME country
- DataError-0031A: SCALE.TRAITID is not equal to FACTOR.TRAITID (given the same SCALEID)
- DataError-0031B: TMETHOD.TRAITID is not equal to FACTOR.TRAITID (given the same TMETHID)
- DataError-0031C: SCALE.TRAITID is not equal to VARIATE.TRAITID (given the same SCALEID)
- DataError-0031D: TMETHOD.TRAITID is not equal to VARIATE.TRAITID (given the same TMETHID)
- DataError-0032A: Level inconsistency in labels of factors in LEVEL_N table
- DataError-0032B: Level inconsistency in labels of factors in LEVEL_C table
- DataError-0034A: Germplasm with METHOD = IMPORT but with no germplasm creation date
- DataError-0034B: Germplasm with incorrect creation date (GDATE) format
- DataError-0034C: Germplasm with creation date (GDATE) after current date
- DataError-0035A: Germplasm with incorrect name date (NDATE) format
- DataError-0035B: Germplasm with NAME date (NDATE) after current date
- DataError-0036A: Genebank accessions (name type GACC) with MTYPE <> Maintenance (MAN)
- DataError-0036B: Genebank accessions (name type GACC) with MGID = 0
- DataError-0036C: Genebank accessions (name type GACC) with GPID1 <> MGID.
- DataError-0036D: Genebank accessions (name type GACC) with GPID1 <> MGID'S GPID1
- DataError-0036E: International Rice Genebank Collection (IRGC) accession with Cross Name (NTYPE = 2)
- DataError-0037: GID in DMS was replaced in GMS
- DataError-0038: Factorid ID or Level No is zero in OINDEX table
- DataError-0039-IRTP/IRGC: IRTP/IRGC accession with invalid characters in names
- DataError-0040A: Duplicate OUNITID-VARIATID IN DATA_C (same value)
- DataError-0040B: Duplicate OUNITID-VARIATID IN DATA_C (different value)
- DataError-0041A: Duplicate LABELID-LEVELNO IN LEVEL_C
- DataError-0041B: Duplicate LABELID-LEVELNO IN LEVEL_N
What's New in Version 5.5.1
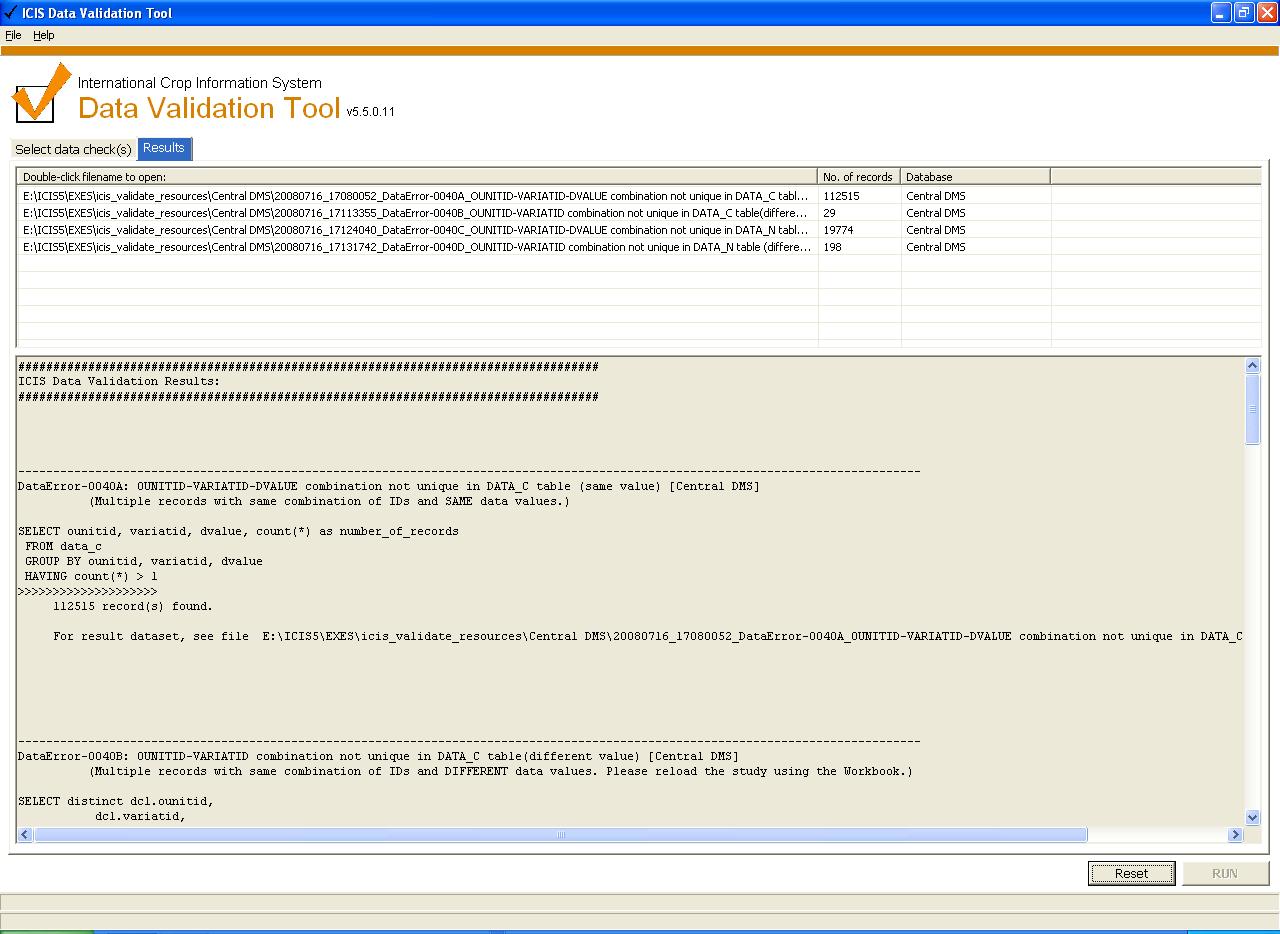
- Improved user interface (display of results):
- 1. Listview that contains the file name, number of records returned, source database. If listview is double-clicked, the specified file will be opened.
- 2. Separate memobox for displaying status messages, SQLs.
- Added option to check local database also.
- New checks for germplasm creation date (GDATE)
- (If germplasm is imported, GDATE should not be blank)
- (Incorrect GDATE format)
- (GDATE later than current date)
- New checks for GMS.NAMES table
- (Name date (NDATE) earlier than germplasm creation date (GDATE)) CropForge Feature Request #631
- (Incorrect NDATE format)
- (NDATE later than current date)
- No NAMES record
- Duplicate names
- New checks for Genebank accession (NTYPE=GACC)
- (MTYPE <> Management (MAN))
- (MGID = 0)
- (GPID1 <> MGID: If a germplasm has an MGID with GPID1=0, then a GID's GPID1 and MGID must be equal )
- (GPID1 <> MGID'S GPID1: If a germplasm has an MGID with GPID1 not 0, then GPID1 and the MGID's GPID1 must be equal )
- New check for DMS: Trait-Scale-Method relationship
- SCALE.TRAITID is not equal to FACTOR.TRAITID (given the same SCALEID)
- TMETHOD.TRAITID is not equal to FACTOR.TRAITID (given the same TMETHID)
- SCALE.TRAITID is not equal to VARIATE.TRAITID (given the same SCALEID)
- TMETHOD.TRAITID is not equal to VARIATE.TRAITID (given the same TMETHID)
- Option to specify/change INI file to use
- Changed error code messaging from "Error-xxxx" to "DataError-xxxx" (prefixed with "Data" to distinguish from error codes of Installation Diagnostic Tool)
- Separation of test status messages and result datasets
- Option to print result dataset(s) in a TEXT file or EXCEL file.
- Option to choose format of filename containing result datasets
- 1. Datetime stamp first or Dataerror code first?
- 2. Include description of DataError in filename?
- If "Print result dataset to MS Excel format" is enabled but no. of records > 65530 (maximum row capacity of Excel is 65536), result dataset will instead be output in a text file.
Data Validation Tool Changes.txt
// // Legend: [+] Addition/New feature // [-] Removal // [*] Modification/Bug Fix // ###################################################################### # # DATA VALIDATION TOOL V5.5.1.1 # ###################################################################### [+] added check for GLOCN of imported germplasms ###################################################################### # # DATA VALIDATION TOOL V5.5.0.11 / V5.5.1.0 OFFICIAL RELEASE # ###################################################################### [+] Added option to specify level of precision in checking of dates (CropForge feature request # 1066) [+] Added grouping of output files in folders according to database (Cropforge feature request # 1058) [+] Addded check for replaced GIDS in DMS (Cropforge feature request #935) [*] Modified SQL for NDATE vs GDATE check (Cropforge feature request #631) ###################################################################### # # DATA VALIDATION TOOL V5.5.0.10 # ###################################################################### [*] Bug fix: Erroneous SQL in check 0023B and 0037E (Cropforge bug # #983) ###################################################################### # # DATA VALIDATION TOOL V5.5.0.9 # ###################################################################### [+] CropForge Feature Request #844: Naming conventions for Data Validation Tool Excel files [+] CropForge feature request #610: Checking the consistency of the Trait, Scale and Method [+] CropForge feature request #710: Check for duplicate names [+] CropFroge feature request #779: Include USERID in reporting of errors [+] Added viewing pane for text/Excel files generated from test [*] Disabled "query with batch update" (temporary only) ###################################################################### # # DATA VALIDATION TOOL V5.5.0.8 # ###################################################################### [*] Modified SQL: checking of foreign key references -- added filter (exclude 0's) [*] Modified SQL: GDATE & NDATE after current date -- added filter (GDATE <> 0; length should be 8) [*] Modified SQL: checking for number of preferred IDs and preferred names [*] Bug fix: preferred ID-name type matching ###################################################################### # # DATA VALIDATION TOOL V5.5.0.7 # ###################################################################### [*] Bug fix: SQL for DataError-0006B (Invalid methods) [*] Modified SQL for checking of progenitor GDATEs, NDATE vs GDATE. ###################################################################### # # DATA VALIDATION TOOL V5.5.0.6 # ###################################################################### [*] Result datasets printed separately from test status messsages. [*] New radiobuttons: Option to print result dataset(s) in TEXT file or EXCEL file [+] Added filter to SQLs: with GERMPLSM.GRPLCE = 0 [*] Bug fix: SQL for DataError-0030 (GID-RELNM-COUNTRY check) ###################################################################### # # DATA VALIDATION TOOL V5.5.0.5 # ###################################################################### [+] New buttons: Select all, deselect all [+] Default printing of result dataset is now on a separate text (.TXT) file. In cases where "Output to Excel" is selected, but no. of records is greater than 65,530: dataset is printed on a text file instead.