Loading of DArT Data into ICIS
From ICISWiki
Contents |
Introduction
Definitions
DArT(Diversity Array technology) is a generic and cost-effective genotyping technology. It was invented by Dr Andrzej Kilian , to overcome some of the limitations of other molecular marker technologies such as RFLP, AFLP and SSR1
P is a cluster variance as a percentage of the total variance of the relative hybridisation intensity of a clone.
PIC is the Polymorphism Information Content of a marker for the set of samples typed.
Call Rate is the percentage of the calls of a clone that are missing.
Discordance
Hamming Distance
Sample DArT data
Dataset Description
Row Headings
- Rows 1-3 are input data to track the samples
- Row 1 is human-readable translation of the barcode affixed to the plate with DNA samples upon arrival
- Rows 2 and 3 are the column (letter) and row (number) of the 96 wells plate that the sample was pipetted from.
- Row 4 has the output headings
Column Headings
- DArT marker name where w signifies wheat, P signifies PstI and t signifies TaqI, specifying the library the clone came from,followed by a numeric marker number.
- DArT clone ID (numeric)
- DArT clone name
- chromosome name to which the marker has been mapped.Chromosome name is only available for those species for which some genetic maps have been constructed. There will be multiple chromosome assignments for a limited fraction of markers that map to more than a single locus.
- P, measuring the quality of the DArT signal for that particular sample
- CallRate
- PIC
- samples which are either known varieties or breeder's lines.
Results
- 0 marker not present in sample
- 1 marker is present in sample
- x missing data
Storing Data in DMS and GEMS
Data stored in DMS
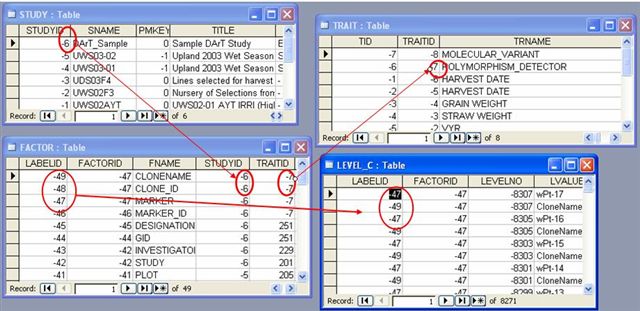
STUDY, FACTOR and TRAIT tables
Loading the sample DArT Data Set into DMS tables will result to the figure below. Each Marker is assigned a Marker ID which is retrieved from the GEMS database. The Marker ID is stored as Factor in the Factor table while MarkerName, CloneID, CloneName are stored as labels of the MarkerID Factor. The MarkerID Factor has the trait "Polymorphism Detector".
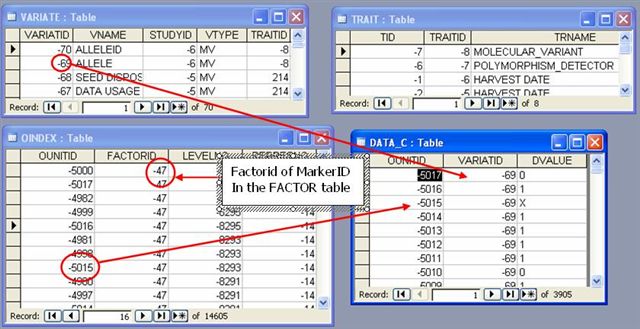
VARIATE, DATA and OINDEX TABLES
Allele or Molecular Variant is also assigned with IDs (ALLELE ID) and is stored as variates of the Marker ID factor in the VARIATES table.
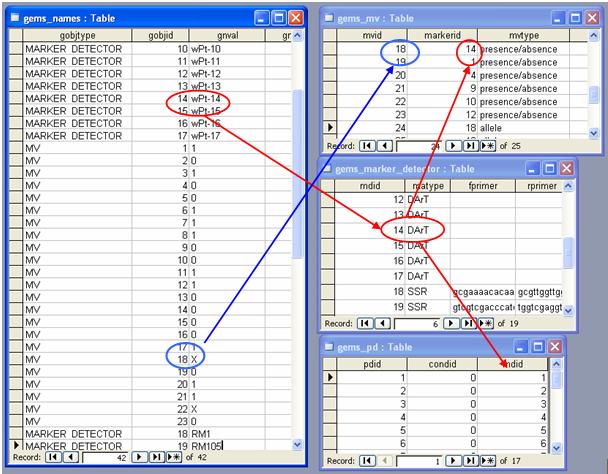
Data stored in GEMS
The MarkerNames from DArT data are stored in the gnval field of the gems_names table. A unique ID (gnid) in the gems_names table is assigned to the MarkerName. It is connected to the gems_marker_detector table thru gobjid if the gobjtype field has the value "MARKER_DETECTOR". The gems_marker_detector is connected to gems_pd table thru the mdid field. The pdid field defines the combination of marker_detector and condition/protocol used for the marker_detector. For markers with more than one protocol, multiple pdids are generated.
Unique IDs are assigned to Allele/Molecular Variant of each Marker and is stored in the gems_names table as gobjid with gobjtype equal to "MV" and is also stored in the gems_mv table as mvid. The gems_mv table contains the information on the Molecular Variant and is connected to the gems_marker_detector table
Loading of DArT Data using ICIS Workbook
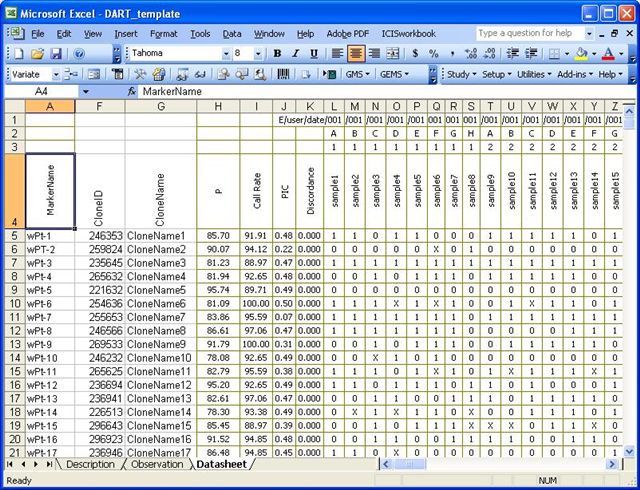
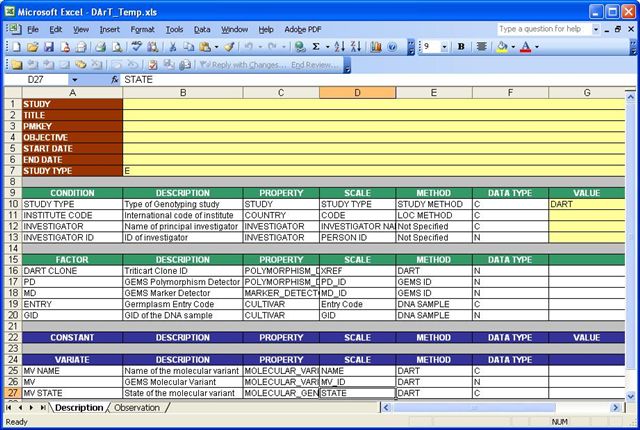
Workbook Template for DArT Data
Workbook Template for DArT Data
DArT Datasheet
The Datasheet below is currently the format that is being delivered to clients. Some DArT formats may include other columns such as discordance and chromosome. This Datasheet is transformed in the Observation format.
Description Sheet
In the Description Sheet, dart clone,PD(Polymorphism Detector), MD(Marker Detector),entry and GID(or samples) are treated as factors. While cloneid, clonename and marker are treated as Labels of the factor markerid. MV, MVNAME and MV STATE are treated as variates. Information about the Study/experiment is also stored in the Description Sheet.
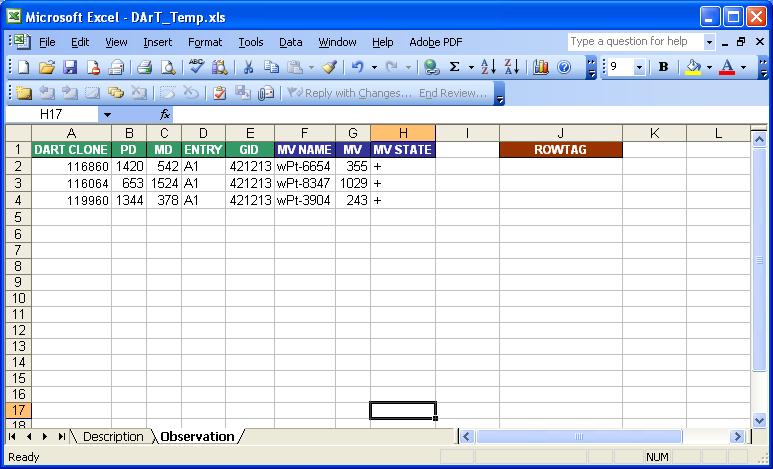
Observation Sheet
The Observation Sheet is created from the Datasheet using a new tool in Workbook for importing Genotype Data. The Observation Sheet is a serialized format of the Datasheet.
The MD(Marker) and MV(Allele) IDs are retrieved from the GEMS database using the Get Marker ID and Get Allele ID tool of the ICIS Workbook. If the Marker or Allele is not yet in the Database, it will be added to the database and a new markerid/alleleid will be assigned to that marker/allele.
Loading of Large DArT datasets
Loading Summary/Derived Data
Derived or Summary data such as PIC, Call Rate and P values are stored as a separate workbook with the same study name. A separate description sheet and Observation sheet will be used to load this data into ICIS.