TDM GMS Overview 5.5
From ICISWiki
THE GENEALOGY MANAGEMENT SYSTEM
INTRODUCTION
The ICIS project seeks to develop a single data management model which can be applied to a wide variety of crops. This model manages information on genealogy, nomenclature, evaluation and characterization of germplasm for a specific crop. The central idea behind the model is that of unique identification of germplasm and management of the homonyms and synonyms which arise naturally in the germplasm development and utilization process. ICIS comprises several modules dedicated to different aspects of the data management problem.
The core of ICIS is a common genealogical data model called the Genealogy Management System (GMS) which must be sufficiently well designed and universal to accommodate a wide range of crops.
The functions of GMS are to:
| a) | assign and maintain unique germplasm identification, | |
| b) | retain and manage information on genealogy and to | |
| c) | manage nomenclature and chronology of germplasm development. |
The genealogical core links to various applications required by individual users. Some of the applications will add genealogical data to the GMS, while others will be analytical, for example, displaying genealogies or calculating coefficients of parentage.
Unique Identification of Germplasm
Since ICIS is an international information system, applications which build or capture genealogical data need to be able to operate independently from site to site and remotely from the central GMS. This presents problems of uniqueness of germplasm identifiers: two remote users must not assign the same identifiers to different germplasm. This problem is resolved by defining a private, or user, GMS for each remote user. The unique identifier is composed of a crop code, followed by a curated user identification number and then by a locally assigned germplasm identifier. For example the rice variety APO has unique identifier IRIS-12-1426 because it was the 1426th germplasm in the local rice database of user number 12. APO now has central GID 204538, but the unique identifier is always kept and never re-used. The central GID has a one-to-one relationship with the unique identifier and so is also unique.
LOGICAL STRUCTURE OF THE GENEALOGY MANAGEMENT SYSTEM
In GMS, all specific instances of germplasm are identified by a unique number called the GERMPLASM_ID (GID). Specific instances of germplasm can be thought of as equivalent to samples of seed which exist or existed at some time, produced by some process or germplasm genesis. In GMS these processes are defined as methods and pertinent information on the genesis of the specific instances are linked to the names via the GID. These methods are classified as those that increase, maintain or reduce genetic variability and are termed generative, maintenance or derivative methods, respectively. The genetic consequences of these methods depend on the reproductive behavior of the species (Table 2.2.a), the genetic structure of the populations which are being manipulated and the breeding strategies being used to achieve genetic change (Annex 2.2.a).
The most common generative processes are the results of crossing individuals or populations and all methods can be included in a general model for crossing practices in plants (Table 2.2.b). The bases for the classification contained in this model is that the source of the female gametes is determined by the source of the seed, which can be seed from a single plant, a selected set of plants or a random set of plants. The source of the pollen needs to be determined but can be from the same plant, another plant, a selected set of plants or random set of plants. The generated germplasm frequently combines material from different sources and as such tends to increase genetic variation or diversity. Information linking these instances of germplasm to their progenitors is retained in the GMS, as well as information on the method of generation of the germplasm.
| Table 2.2.a: Reproductive Systems in Crop Plants | |
| Self Pollinated1 | Inbreeding, self pollinated or self fertilised species. Examples: Wheat, Rice, Barley |
| Predominantly Self Pollinated2 | Examples: Cotton, Pigeonpea, Sorghum |
| Open Pollinated1 | Outbreeding, cross pollinated or cross fertilised species. Examples: Maize, Pearl Millet, Cucurbits |
| Self Incompatible3 | Self incompatible species. Examples: Rye, White Clover, Papaya |
| Vegetatively Propagated4 | Vegetatively or clonally propagated species. Examples: Potatoes, Casava, Yam, Taro, Sugar Cane, Pineapple, Strawberries |
| Apomictic5 | Apomitic (produce seed by asexual means) species. Examples: Green Panic, Buffel grass |
1 Catered for in the ICIS Methods Table.
2 Probably catered for as breeding methods usually follow those for Self Pollinated crops.
3 Probably catered for by methods suitable for Open Pollinated crops.
4 Generative and derivative and most maintenance methods catered for as these crops will be either self or cross fertilised in their mode of sexual reproduction.
5 Probably catered for by methods suitable for Vegetatively Propagated crops
Other items of germplasm arise by derivative processes from a single source. Derivative methods are divided into two categories, derivative germplasm development methods (DER) which tend to refine and reduce genetic variation and germplasm management methods (MAN) which attempt to preserve variation at the level of the source. Common derivative processes are selection and seed increase. Seed increase attempts to maintain genetic variability whereas selection tends to refine and reduce genetic variation. Both lead to instances of germplasm which are closely related both to the source and to different derivations from a common source. All instances of germplasm derived from a single source form a related group (or “family”) of germplasm. Information on the method of derivation of such germplasm is also stored. Management methods attempt to maintain genetic variation and leads to instances of germplasm which are closely related, preferably identical genetically, to the common source.
A full list of methods classified by breeding method of the crops are given in Annex 2.2.a and a full description and listing of the METHODS TABLE are given in Annex 2.2.b. The methods supplied in GMS are a compromise between giving a parsimonious set derived strictly from the model given in Table 2.2.b and an attempt to define every possible method that has or could be used. The first option does not provide enough information to crop coordinators, local coordinators or users. The second swamps users with too much information and a bewildering array of choices. Providing users with the full menu of methods provided in Annex 2.2.b would be counter productive (there are too many producing confusion) and ICIS will enable crop and local coordinators to select a subset for the crop or project that the service. A warning screen should be displayed asking users to explore already defined protocols and adding these to their menu before defining their own.
| Table 2.2.b: Schematic outline of a general model for crossing practices in plants. | |||
| The bases for the classification contained in this model is that the source of the female gametes is determined by the source of the seed, which can be a single seed or seed bulked from a single plant, a selected set of plants or a random set of plants. The source of the pollen needs to be determined but can be from the same plant, another plant, a selected set of plants or a random set of plants. The genetic consequences of these crosses or selections depends on the reproductive behaviour of the species. | |||
| Female Source | Male Source | Generative Methods | Derivative Methods |
| Single Plant | |||
| Same Single Plant | 40(G) Selfing | 208(S), 505(O) Single Seed Descent 60(G) Plant Identification 205(S), 504(O) Single Plant Selection 209(S), 506(O) Restorer Selection 210(S), Maintainer Selection | |
| Different Single Plant | 101(S),401(O) Single Cross 102(S),402(O) Three-way Cross 103(I),403(O) Double Cross 404(O) Full diallel cross 405(O) Full diallel cross bulked 406(O) Half diallel cross 407(O) Half diallel cross bulked 408(O) Partial diallel cross 409(O) Partial diallel cross bulked 424(O) Convergent Cross 425(O) Partial Plus Diallel Cross 107(S) Backcross 108(S) Backcross Recessive 109(S) Interspecific Cross 416(O) Narrow based tester, line CF | ||
| Selected Bulk | 104(S) Female Complex Top Cross 110(S) Selected Pollen Cross SF 416(O) Narrow based tester, line CF 418(O) Broad based tester, Line CF 417(O) Narrow Based Tester, POP CF | ||
| Bulk | 104(S) Female Complex Top Cross 111(S) Random Pollen Cross SF 426(O) Random Half Sib Families 427(O) Selected Half Sib Families | ||
| Selected Bulk | |||
| Single Plant | 105(S) Male Complex Top Cross 108(S) Backcross Recessive 416(O) Narrow based tester line CF | ||
| Same Selected Bulk | 420(O) Polycross CF 421(O) Random Mating CF | 203(S) Purification 204(S), 503(O) Rouging 206(S) Selected Bulk 508(O) Full Mass Selection | |
| Different Selected Bulk | 106(S) Complex Cross 112(S) Open Pollenated SF 404(O) Full diallel cross 405(O) Full diallel cross bulked 406(O) Half diallel cross 407(O) Half diallel cross bulked 408(O) Partial diallel cross 409(O) Partial diallel cross bulked 410(O) Subset Cross 411(O) Population backcross CF 412(O) Selected Pollen Cross CF 413(O) Interspecific cross CF 417(O) Narrow Based Tester, POP CF 419(O) Broad Based Tester, POP CF 423(O) Population Cross CF | ||
| Bulk | 415(O) Random Pollen Cross CF | 509(O) Half Mass Selection | |
| Bulk | |||
| Single Plant | 105(S) Male Complex Top Cross 411(O) Population Backcross CF 412(O) Population Backcross Recessive CF | ||
| Selected Bulk | 411(O) Population Backcross CF 412(O) Population Backcross Recessive CF 414(O) Selected Pollen Cross CF 419(O) Broad Based Tester, POP CF 423(O) Population Cross CF | ||
| Same Bulk | 422(O) Open pollenation CF | 207(S), 507(O) Random Bulk | |
| Different Bulk | 106(S) Complex Cross 112(S) Open Pollenation SF 413(O) Interspecific Cross CF 415(O) Random Pollen Cross CF 422(O) Open pollenation CF 423(O) Population Cross CF | ||
| I: Inbreeding = S: Self Breeding = Self Fertilisation O: Outbreeding = C: Cross Breeding = C: Cross Fertilisation | |||
An example of the compromise occurs between the export and import protocols and the acquisition, seed increase and cultivar generation protocols. Only a generic method is given for the import and export methods (methods 62 and 63) but a series of detailed methods specifying the type of material acquired, increased or released as cultivars are given for the later protocols. It is expected that these decisions will be tested as the number of crops included in ICIS increases. The import and export protocols will connect germplasm to its genealogy if the source is already in ICIS. If not a problem will arise if the source is entered either through another local user or by entry from a program that is independent from ICIS. Never the less, ICIS provides a mechanisms for users to define new methods for germplasm generation, management or derivation.
There are no conventions which are universally applied to the naming of breeding methods, strategies, or genetical material used in plant breeding programs. For example, released germplasm from plant breeding programs are referred to as ‘varieties’ or ‘cultivars’ or rarely as ‘cultigens’. Both the first and last are incorrect as ‘variety’ is a taxonomic term and no released genetic material consists of a single genotype. ICIS uses ‘cultivar’ as the generic definition for released genetic material for agricultural use.
The genetic consequences of these crosses or selections depends on the reproductive behavior of the species. For example re selection or collection from a landrace population or transgenic generation could link back to one or no progenitor, while complicated crosses involving, for example, multiple pollen parents could link to many. Similar methods for different crops have different genetic consequences depending on the reproductive system of the crop, self-pollinated, cross-pollinated or clonally propagated. These are distinguished in GMS as separate method groups (MGRP) in the methods table (Annex 2.2.b) as S, O, C respectively or G for general methods which apply to all crops.
Germplasm Information
Information of the generation or derivation of any instance of germplasm is stored in the following variables:
| Table 2.2.1.a: Germplasm Information stored in GMS. | |
| GERMPLASM_ID | Germplasm Identification Code. This is unique for any germplasm recorded. The general rule is: if you wouldn’t mix the packets of seed that exist or did exist you give the GEN an entry in GMS. |
| METHOD_NO | This is the method of genesis of germplasm and details are stored in the method table (Annex 2.2.b). The method indicates whether the germplasm was produced by a generative, derivative or preservative process, and then how the germplasm was produced. Details of the method which change with each application are stored as attribute information (Section 2.2.4). |
| GROUP_ID | Identification code for last instance of germplasm produced by a generative process from which the current instance was derived. GROUP_ID = GERMPLASM_ID if the current instance is produced by a generative process. |
| SOURCE_ID | Identification code for the immediate source of a derived or preserved germplasm. (Not valid for generated germplasm.) |
| GERMPLASM_USER_ID | Identification code of the user who submitted data on the genesis of the germplasm. |
| LOCATION_NO | Identifier of breeding or origin location. |
| GERMPLASM_DATE | Date of generation, derivation or preservation of the germplasm. |
| GERMPLASM_REFERENCE | Identifier of bibliographic reference from where the germplasm data was retrieved. |
Link Information
Link information is information on the links between an instance of generated germplasm and its progenitors. The number of progenitors and the role of each, such as male or female, is defined by the METHOD in the germplasm information. The link variables required for each progenitor are:
| Table 2.2.2.a: Link information stored in GMS. | |
| GERMPLASM_ID | Germplasm Identification Code. |
| NO_PROGENITORS | Number of progenitors. |
| PROGENITOR_ID | Progenitor GERMPLASM_ID. |
Name Information
A third form of information retained by GMS manages nomenclature and chronology of names of germplasm. Names are any designation used to identify germplasm and this includes breeders’ IDs, release names, abbreviations and accession numbers. The required variables are:
| Table 2.2.3.a: Name information stored in GMS. | |
| GERMPLASM_ID | Germplasm Identification Code. |
| NAME_TYPE | Some names can belong to several instances of germplasm, some to only one, and some instances can have several names of the same type. These relations are indicated by NAME-TYPE. For example some types would be: ACCNO - Accession No in a germplasm bank (USER_ID points to the germplasm bank as a user of the system) BCID - Breeder’s Cross ID CVNAM - Release or Cultivar Name BRNAMBreeder’s Name, a cross name and selection history in the breeder’s own notation ALNAM - Breeder’s advanced line name. Usually assigned when BRNAM gets too long. NTNAM - National testing name PONAM - Population name CNNAM - Cycle name for recurrent selection schemes SYNAM - Synthetic cultivar name HYNAM - Hybrid cultivar name ADVNM - Alternative derivative name ACVNM - Alternative cultivar name AABBR - Alternative abbreviation Note: Provision is made for a user to add to these names. |
| NAME_STATUS | Number indicating the storage type and status of the name: 0 - ASCII names which are alternative names. 1 - ASCII names which are preferred names. 2 - ASCII names which are preferred abbreviations. 3 - Chinese-GBK (GD) DBCS names. 4 - Chinese Big 5. 5 - Japanese. 6 - Korean. 8 - ASCII names which are preferred IDs. 10 - UNICODE names which are not preferred. Note: The preferred name must be in ASCII. |
| NAME_USER_ID | The source of name information. This is important for integrity, referencing and elimination of homonyms. The user registration number is sufficient since only registered users can provide information to the system. This deals with the problem of homonyms; by incorporating the USER_ID as part of the germplasm name, since a single user would not assign the same name to more than one piece of germplasm in the local user’s system |
| NAME_VALUE | Name value assigned by a user to a specific instance of germplasm. |
| NAME_LOCATION_NO | Location ID where the name was assigned |
| NAME_DATE | Date of name recording or assigning |
| NAME_REFERENCE | Identifier of bibliographic reference from where the name data was retrieved. |
Attribute Information
A fourth form of information retained by GMS records any attributes of the germplasm the user would like to retain. These attributes should be controlled by the central database manager so that only attributes relating to genealogy and nomenclature are stored in GMS. The required variables have an exactly similar structure to the names variables as follows:
| Table 2.2.4.a: Attribute Information stored in GMS. | |
| GERMPLASM_ID | Germplasm Identification Code. |
| ATTRIBUTE_TYPE | Some attributes can belong to several instances of germplasm, some to only one, and some can have several values for a single instance of germplasm. These relations are indicated by ATTRIBUTE_TYPE. For example some types would be: BSTRBreeder’s Selection String (These BSIDs note the specific selection step in a selection history in the breeders own notation. The full selection history may also be stored as a line name in the NAMES table. SPECSpecies OSETOriginating set. The germplasm set from the set generation module where the germplasm was first derived. (See set generation later.) |
| ATTRIBUTE_USER_ID | The source of attribute information. |
| ATTRIBUTE_VALUE | Attribute value assigned by a user to a specific instance of germplasm. |
| ATTRIBUTE_LOCATION_NO | Location of attribute determination. |
| ATTRIBUTE_DATE | Date of attribute determination. |
| ATTRIBUTE_REFERENCE | Identifier of bibliographic reference from where the attribute data was retrieved. |
The following are definitions of attributes which add information about the methods are stored in the USER_DEFINED_FIELDS table.
| Table 2.2.4.b: Definitions of attributes which add information about methods. | ||
| FLDNO | FCODE | FNAME |
| 103 | NOTES | Notes concerning germplasm origin, development or chronology. |
| 104 | RELEASE | Cultivar type. |
| 208 | BS | Number of plants bulked and target traits or genes. |
| 209 | IC | Target introgression in interspecific cross |
| 210 | MT | Agent and method of induced mutation. |
| 211 | MATT | A description of the method or agent used for a specific instance of the method. |
| 212 | RMA | Attributes for random mating methods. These are: NPSnumber of parents randomly mated. PCTpercent contribution from each parent. MMTHintervention method to |
| 213 | MCOLL | Methods of collection. |
| 214 | IMPORT | Type of germplasm. See Table 2.2.a for lists of types of germplasm. |
| 215 | BLKM | Method of bulking seed, possibilities are: Bulking all seed from plants. Bulking equal volume from each plant. Bulking equal weight from each plant Bulking equal number from each plant |
| 216 | SUBSETA | No of subsets and bulking method. See above for bulking methods. |
| 221 | ST | Target traits or genes. |
| 222 | SEX | Selfs excluded, Yes or No |
EXAMPLES OF STORAGE OF GENEALOGY INFORMATION
Several examples of storage of genealogy information are given in this section. Examples of the type of information generated by the system are given and the entry of genealogical information for the rice cultivar IR24 is given as an example of storing historical data. The examples used to illustrate system storage include a modified pedigree plant breeding system in wheat, an open recurrent selection system in wheat, a reciprocal recurrent selection system in maize, a polycross among potato clones and a half diallele using the same four clones.
Genealogy Management for a Modified Pedigree Breeding Program
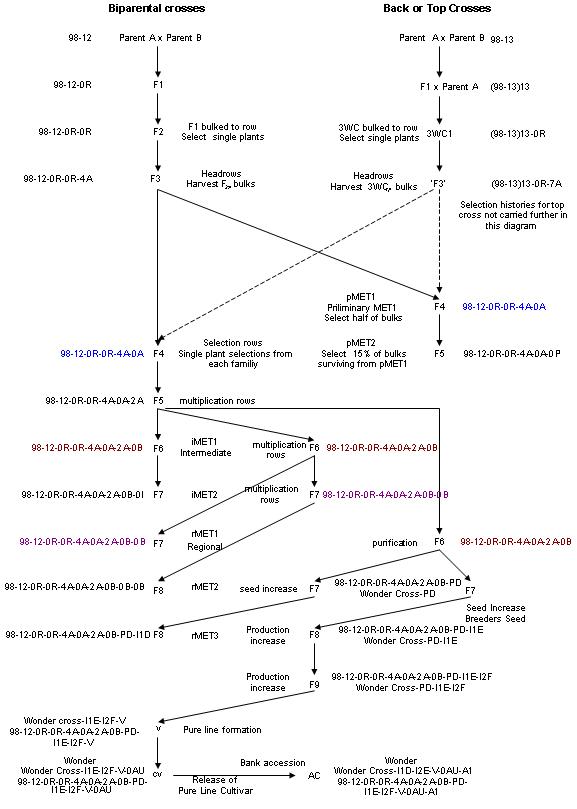
The modified pedigree breeding program illustrated here (Figure 2.3.1.a ) is modified from the program used by the wheat breeding program at the Leslie Research Centre of the Queensland Department of Primary Industries in Australia. This example illustrates the use of the three types of methods used in GMS, generative, derivative and maintenance methods. The system used for the selection history (Figure 2.3.1.a) and used as a breeders name (Table 2.3.1.a) is a modification of the that used by the CIMMYT wheat program and is for illustrative purposes only. It is not the system used by the LRC program.
Generative methods
The example provides an illustration of two common generative methods used in pedigree breeding programs, viz. a two way and a three way cross. The three-way cross in this example is a top-cross as the F1 from the first cross is crossed to another adaptive line.
For the two way cross assume that line A (GID821; table row 1) is the female parent (FPARNT) and line B (GID822; table row 2) is the male parent (MPARNT) of the single cross (generative method (METH C2W)) 98 12 (GID900; table row 4). 98 12 is the 12th cross made in 1998 and has two progenitors (No PROGEN). Note that all lines listed in the table which are derivatives of this two way cross have the pedigree A/B but have different selection histories.
For the three-way cross the F1 (GID900, row 4) from the two-way cross, used as a female, is mated (in the next year) to another adapted parent C (GID823, table row 3) to produce the three-way cross (3WC) (GID950, table row 5) and this is the 13th cross made in 1999. All derivatives of this cross have the pedigree A/B//C. Note that while this pedigree does not indicate whether A/B was an F1 or a fixed line this information is carried in the GERMPLASM TABLE. This clearly indicates that GMS does not store pedigrees, but rather genealogies. Purdy pedigrees are an output (query) on GMS and other forms of a ‘pedigree’ can be programmed as a query.
Derivative methods
The F1 is a bulk of all the grains produced from the cross (table row 7, GID1001, METH DRB) and is designated 98-12-0R where 0R indicates a bulk and R indicates the place where the F1 was grown (Research Station in this example). Note that the F1 (and all further derivatives) has as a group number the GID (900) of the cross from which they were derived. The source number is the GID of the immediate packet of seed from which each line was derived, in this case the cross GID. The F2 (table row 9, GID2001, METH DRB) is a random bulk of the F1 and is designated 98-12-0R-0R as in this example the F1 was grown on the research station. It has group number GID900 and source number GID1001. The F2 rows are grown at place A. The F2 is segregating and single plant selections are taken and these F3 lines (table rows 11 & 12 are examples) are grown as ‘head rows’ also at place A. As these are single plant selections, the resultant F3 lines are designated 98-12-0R-0R-4A (table row 11) etc where 4A represents the fourth plant selected from this F2 at place A. These F3 lines are again grown as head rows at place A and bulked. The resultant F2 derived F4 bulks are designated 98-12-0R-0R-4A-0A (table row 15) and are used to sow unreplicated preliminary yield trials (designated P in this example) and to grow rows for single plant selections at place A.
In the CIMMYT system for writing selection histories, and in most if not all other systems including IRRI, the two steps indicating the F1 and F2 are omitted in the selection histories for single crosses as the first two -0R-0R- steps are common to all derivatives of a single cross. Using this system the F3 lines would be designated 98-12-4A indicating that this the 4th single plant selection from the F2 of the cross 98-12.
The 3WC1 (table row 8, GID1002, METH DRB) designated (98-13)13-0R is a bulk of all seeds from the three-way cross, GID950 and is grown as a row at place A. It has the group number GID950 of the cross from which it comes and the same GID as its source number. Unlike the F1, which is homogeneous, this population is segregating. Hence single plant selections are often taken from these population types rather than a bulk as is the case for the F1 from a single cross. These ‘F3’ single plant selections are grown as head rows at place A and because they are single plant selections they are designated (93-13)13-0R-7A (row 13) for example. This line has GID950 as its group code and GID2010 as its source code. These lines are treated from this point on in the same way as the single crosses.
In the three-way cross only the 1st -0R- step is common to all derivatives and the commonly used (CIMMYT/IRRI) system would designate the ‘F3’ line as (98-13)13-G indicating that this was the 7th single plant selection from the bulk of all the seed derived from the three-way cross (98-13)13.
Figure 2.3. 1 Diagram of a stylised representation of a modified pedigree plant breeding program.
The F4 bulks (table rows 15 & 16) are grown in preliminary multi-environment trials (pMET1) for one year.
F5 bulks (eg table row 17, GID2201, METH DRB) from those selected from pMET1 (50%) are grown for a second year in pMET2 to enable two years of data to be used for selection for yield among the F2 derived F4 bulks.
The surviving F4 lines selected using data from the pMETs are grown as rows at site A and single plant selections made.
The F5 lines derived (eg table rows 19, GID2301, designated 98-12-0R-0R-4A-0A-2A) are planted as seed multiplication rows at site B.
The resultant F6 bulks (eg table row 21, GID3001, METH DRB, designated 98-12-0R-0R-4A-0A-2A-0B) are sown in the first year of a replicated intermediate MET (iMET1), sown at site B for seed multiplication for the first year of regional METs (rMETs) and re sown at site B in the first year of rMETs for seed purification if the line looks promising for release. Once a line is tested in a MET it acquires a breeders name (BN21 and BN22) in these cases.
The F7 bulk seed from iMET1 (eg table row 23, GID3101, 98-12-0R-0R-4A-0A-2A-0B-0I) is used for iMET2. The addendum letter I is used here to designate seed produced from an intermediate MET.
The F7 bulk seed produced from the multiplication rows (table rows 25 & 26) at site B is used for the first regional MET (rMET1) and for a further multiplication of seed, at site B, to produce the F8 bulk seed for rMET2 (table rows 27 & 28).
Maintenance methods
The example from hear on will follow the history of an F6 line, GID3001, which goes on to be released as a cultivar. This process consists of one derivative method, seed purification, and the maintenance methods, seed increase for yield testing in the final year of the rMETs, a seed multiplication to produce breeders seed for release of a cultivar called in this example Wonder. At this stage if more than one line from the cross is used as a parent the cross may be named, here Wonder Cross. The process of release consists of two production seed increases to obtain enough seed for release and the release process itself.
Table row 29 (GID4001, METH DPU) represents the purification at site B of BN21 by derivative method seed purification to produce F7 pure seed. Note that I have suggested the addendum -PD to indicate purification at site D. The F7 pure seed is used for a seed increase at site B to produce F8 seed for testing in the final rMET3 and for producing breeders seed at site E.
Table row 30 (GID4101, METH NBK) represents the maintenance method ’bulk seed increase’ at site B of GID4001 for seed for testing in rMET3.
Table row 31 (GID4201, METH VBS) represents the maintenance method ’breeders seed production’ at site E. An addendum -I1E is used to indicate the first seed increase conducted at site E.
Table row 32 (GID4201, METH NBK) represents the maintenance method ’bulk seed increase’ at site F. This is the second seed increase, hence the addendum -I2F, representing the second seed increase which was done at site F.
Table row 33 (GID4401, METH VPL) represents the maintenance method ’pure line formation’ in preparation for releasing a pure line cultivar. In this case this does not represent a physical process but does in many situations, eg forming and F1 hybrid.
Table row 34 (GID4410, METH VCR) represents the maintenance method ’cultivar release’. This process should be maintained separate from the formation odf a cultivar. It can occur more than once for the same material.
| Table 2.3.1.a: Germplasm Table for Selection Histories for an example Pedigree Breeding Program. | |||||||
| Generative Methods | Breeder’s name | ||||||
| Row in table | GID | METH | No PROGEN | FPARNT | MPARNT | ||
| 1 | 821 | A | |||||
| 2 | 822 | B | |||||
| 3 | 823 | C | |||||
| . | . | . | . | . | . | . | . |
| 4 | 900 | C2W | 2 | 821 | 822 | 98-12 | Single cross |
| 5 | 950 | C3w | 2 | 900 | 823 | (98-12)13 | Three-way cross |
| Derivative or Management Methods | |||||||
| Row in table | GID | METH | GROUP | SOURCE | |||
| 7 | 1001 | DRB | 900 | 900 | 98-12-0R | F1 rows | |
| 8 | 1002 | DRB | 950 | 950 | (98-13)13-0R | 3WC1 rows | |
| . | . | . | . | . | . | . | . |
| 9 | 2001 | DRB | 900 | 1001 | 98-12-0R-0R | F2 rows | |
| 10 | 2010 | DRB | 950 | 1002 | (98-13)13-0R | 3WC1 rows | |
| . | . | . | . | . | . | . | . |
| 11 | 2101 | DSP | 900 | 2001 | 98-12-0R-0R-4A | F3 rows | |
| 12 | 2102 | DSP | 900 | 2001 | 09-12-0R-0R-5A | F3 rows | |
| 13 | 2151 | DSP | 950 | 2010 | (98-13)13-0R-7A | ’F3’ rows | |
| 14 | 2152 | DSP | 950 | 2010 | (98-13)13-0R-8A | ’F3’ rows | |
| . | . | . | . | . | . | . | . |
| 15 | 2171 | DRB | 950 | 2101 | 98-12-0R-0R-4A-0A | F4 bulks | |
| 16 | 2172 | DRB | 950 | 2102 | 98-12-0R-0R-5A-0A | F4 bulks | |
| . | . | . | . | . | . | . | . |
| 17 | 2201 | DRB | 900 | 2171 | 98-12-0R-0R-4A-0A-0P | F5 bulks | |
| 18 | 2202 | DRB | 900 | 2172 | 98-12-0R-0R-5A-0A-0P | F5 bulks | |
| . | . | . | . | . | . | . | . |
| 19 | 2301 | DSP | 900 | 2201 | 98-12-0R-0R-4A-0A-2A | F5 rows | |
| 20 | 2302 | DSP | 900 | 2202 | 98-12-0R-0R-4A-0A-3A | F5 rows | |
| . | . | . | . | . | . | . | . |
| 21 | 3001 | DRB | 900 | 2301 | 98-12-0R-0R-4A-0A-2A-0B | F6 bulks BN21 | |
| 22 | 3002 | DRB | 900 | 2302 | 98-12-0R-0R-4A-0A-3A-0B | F6 bulks BN22 | |
| . | . | . | . | . | . | . | . |
| 23 | 3101 | DRB | 900 | 3001 | 98-12-0R-0R-4A-0A-2A-0B-0I | F7 bulks BN21 | |
| 24 | 3102 | DRB | 900 | 3001 | 98-12-0R-0R-4A-0A-3A-0B-0I | F7 bulks BN22 | |
| . | . | . | . | . | . | . | . |
| 25 | 3201 | DRB | 900 | 2301 | 98-12-0R-0R-4A-0A-2A-0B-0B | F7 bulks BN21 | |
| 26 | 3202 | DRB | 900 | 2302 | 98-12-0R-0R-4A-0A-3A-0B-0B | F7 bulks BN22 | |
| . | . | . | . | . | . | . | . |
| 27 | 3301 | DRB | 900 | 3201 | 98-12-0R-0R-4A-0A-2A-0B-0B-0B | F8 bulks BN21 | |
| 28 | 3302 | DRB | 900 | 3202 | 98-12-0R-0R-4A-0A-3A-0B-0B-0B | BN22 | |
| . | . | . | . | . | . | . | . |
| 29 | 4001 | DPU | 900 | 3001 | 98-12-0R-0R-4A-0A-2A-0B-PD | F7 pure line Wonder cross | |
| 30 | 4101 | NBK | 900 | 4001 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1D | Wonder cross | |
| 31 | 4201 | VBS | 900 | 4101 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1E | Wonder cross | |
| 32 | 4301 | NBK | 900 | 4201 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1E-I2F | Wonder cross | |
| 33 | 4401 | VPL | 900 | 4301 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1E-I2F-V | Wonder cross | |
| 34 | 4410 | VCR | 900 | 4401 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1E-I2F-V-0AU | cv Wonder | |
| 35 | 4501 | ISE | 900 | 4401 | 98-12-0R-0R-4A-0A-2A-0B-PD-I1E-I2F-0AU-AC | cv Wonder WA28 | |
| ID = is unique germplasm identifier. METH = is the breeding method, either generative, derivative or management. No PROGEN = is the number of progenitors of a derivative method. FPARNT = ID of female parent. MPARNT = ID of male parent. GROUP = ID of last generative process for the derivative or management process. SOURCE = ID of immediate predecessor for the derivative or management process. | |||||||
Finally, in this example, the seed is placed in a germplasm bank. This is represented by table row 36 (GID4501, METHH ISE), the ‘seed import’ method and acquires another name WA28.
In this example many of the seed increase operations reflected in the selection histories are carried out at different sites. If these operations are carried out at the same site some modification of the system for naming selection histories may be required to reflect these differences should this be required.
Genealogy Management for An Open Recurrent Selection Method
The recurrent selection method for wheat improvement being used by the University of Queensland in their Germplasm Enhancement Program conducted at the Leslie Research Institute in Australia is described by Fabrizius et al. (1996). This note indicates how pedigrees from that system can be recorded in the GMS structure of ICIS.
- (A) The open reciprocal selection method starts with the selection of a number of parental lines from which a population is developed by randomly mating the F1s from a complete diallel using a gametocyte. Consider a system based on 4 parents with GIDs 871, 927, 933 and 1001 as indicated in rows 1, 2, 5 and 8 of the GERMPLASM TABLE (Table 2.3.2.a.). The 6 F1s from the diallel could be simply entered as 1004 to 1009 as shown on rows 12-17 in Table 2.3.2.a.
- (B) The first step of the recurrent selection cycle is to randomly mate the F1s using a gametocyte. To record this we need to define a method in the METHODS TABLE for this form of random mating. Suppose we use code CGO (Method 112 in Table 2.1.d) for the method which has a variable number of parents (defined for each implementation by the NO_ OF_ PROGENITORS parameter in the GERMPLASM TABLE). Method CGO also has no defined number of female parents since all parents act as male and female parents. In our example there are 6 parents, for the random mating recorded in row 18 of Table 2.3.2.a. The first two parents are recorded in the GERMPLASM TABLE as in row 18, the remaining 4 are recorded in the OTHER PROGENITORS TABLE as in rows 1 to 4 of Table 2.3.2.b. The resulting population is termed the C0 population.
- (C) The next step in the process is to screen a large number of S0 plants from C0 seed for semidwarf status and rust resistance. A number of these are kept and proceed through a generation of seed increase and rust testing. In practice the number kept will be 2,000, but for our example we record the pedigree of 10 in rows 19 to 28 of Table 2.3.2.a. These retained S1 families represent single plant selections, DSP, out of the C0 population.
- (D) Selections are made on the basis of rust scores from the S1 plants, and a number are kept. In practice this number will be 800, but for our example we will consider 5. Seed from these retained families is split for storage, multi-environment testing for two years and bulk quality testing for the survivors. Suppose our retained S1 families are C0-2, C0-3, C0-7, C0-9 and C0-10 from rows 20, 21 25, 27 and 28 of Table 2.3.2.a. These are bulk selected in the third year, with surviving families recorded as A on rows 29 to 33 of Table 2.3.2.a and again in the fourth year, B, to produce four remaining families C0-2AB, C0-7AB, C0-9AB and C0-10AB recorded on rows 34 to 37 of Table 2.3.2.a.
- (E) It is possible, as an off-shoot of the system, that single plant selection can be made from the evaluation year nurseries and be incorporated into traditional breeding programs, for example the third plant from the family C0-2A in row 29, and the sixth from C0-7AB in row 35 could be selected for incorporation in other breeding programs. These are the families recorded in the usual manner as in rows 38 and 39 of Table 2.3.2.a.
- (F) Following quality testing of the families surviving two years of evaluation, a number of families will be selected to go into a second round of random mating. In fact about 100 will be selected, but we will consider 3 of our surviving families C0-2AB, C0-7AB and C0-9AB. The seed for this random mating cycle will however come from the S1 retained seed, C0-2, C0-7 and C0-9 in our example. As a variation on simple random mating new parental lines can be introduced at this point by including plants from new lines in the random mating process at specified proportions of the population. This is the step that gives rise to the term Open Random Mating, and as this is not in the METHODS TABLE (Table 2.1.d) we need to define a new method, ORM, with variable number of parents. This emphasises the point made previously about the necessity of defining all possible methods for the methods table. The methods defined are a compromise between a too parsimonious set which does not convey enough information and attempting to define every possible method and swamping the user with too much information. The compromise allows new methods to be defined by users 9with MIDs over 1000 and accepted or not by the ICIS crop coordinator.
ORM also has no defined female parents, but a variable number of imported parents, and an incorporation percentage for each. These parameters must be stored as attributes of the breeding method for each implementation of the method. In the METHODS TABLE, the ATTRIBUTES flag would be set for method ORM to indicate that attribute values for each implementation are stored in the ATTRIBUTES TABLE. THE CONSTANTS TABLE would define these values as number of imported parents which are specified last in the list of parents, and percent contribution of each. The CONSTANTS TABLE would also indicate that each value occurred on an extension line of the attribute in a specified format. For our example, suppose we wish to retain three S1 families and include two new parents with IDs 930 and 1000 (as in rows 3 and 7 of Table 2.3.2.a) with proportions 15% and 10% of the random mating population.
The new population would be recorded as in row 40 of Table 2.3.2.a with GID 1032. The first two parents, C0-2 and C0-7 are on that line, the remaining three are recorded on the OTHER PARENTS TABLE (rows 5 to 7 in Table 2.3.2.b). The attribute values would be stored on the ATTRIBUTES TABLE as in Table 2.3.2.c. The positive value of ATTRIBUTE-EXT indicates the number of attribute values, in this case one more than the number of imported parents, and the negative values define the order of the remaining values down to -1, the last value.
| Table 2.3.2.a: Germplasm Table For Open Recurrent Selection Example. | ||||||||
| Row in Table | GID | Method | Number of progenitors | Generative Process | Derivative Process | Name | ||
| Female Parent | Male Parent | Group | Source | |||||
| 1 | 871 | |||||||
| 2 | 927 | |||||||
| 3 | 930 | |||||||
| 4 | ||||||||
| 5 | 933 | |||||||
| 6 | ||||||||
| 7 | 1000 | |||||||
| 8 | 1001 | |||||||
| 9 | ||||||||
| 10 | ||||||||
| 11 | ||||||||
| 12 | 1004 | C2W | 2 | 871 | 927 | . | . | Fdn 12 |
| 13 | 1005 | C2W | 2 | 871 | 933 | . | . | Fdn 13 |
| 14 | 1006 | C2W | 2 | 871 | 1001 | . | . | Fdn 14 |
| 15 | 1007 | C2W | 2 | 927 | 933 | . | . | Fdn 23 |
| 16 | 1008 | C2W | 2 | 927 | 1001 | . | . | Fdn 24 |
| 17 | 1009 | C2W | 2 | 933 | 1001 | . | . | Fdn 34 |
| 18 | 1010 | CGO | 6 | 1004 | 1005 | . | . | C0 |
| 19 | 1011 | DSP | 1 | . | . | 1010 | 1010 | C0-1 |
| 20 | 1012 | DSP | 1 | . | . | 1010 | 1010 | C0-2 |
| 21 | 1013 | DSP | 1 | . | . | 1010 | 1010 | C0-3 |
| 22 | 1014 | DSP | 1 | . | . | 1010 | 1010 | C0-4 |
| 23 | 1015 | DSP | 1 | . | . | 1010 | 1010 | C0-5 |
| 24 | 1016 | DSP | 1 | . | . | 1010 | 1010 | C0-6 |
| 25 | 1017 | DSP | 1 | . | . | 1010 | 1010 | C0-7 |
| 26 | 1018 | DSP | 1 | . | . | 1010 | 1010 | C0-8 |
| 27 | 1019 | DSP | 1 | . | . | 1010 | 1010 | C0-9 |
| 28 | 1020 | DSP | 1 | . | . | 1010 | 1010 | C0-10 |
| 29 | 1021 | DRB | 1 | . | . | 1010 | 1012 | C0-2A |
| 30 | 1022 | DRB | 1 | . | . | 1010 | 1013 | C0-3A |
| 31 | 1023 | DRB | 1 | . | . | 1010 | 1017 | C0-7A |
| 32 | 1024 | DRB | 1 | . | . | 1010 | 1019 | C0-9A |
| 33 | 1025 | DRB | 1 | . | . | 1010 | 1020 | C0-10A |
| 34 | 1026 | DRB | 1 | . | . | 1010 | 1021 | C0-2AB |
| 35 | 1027 | DRB | 1 | . | . | 1010 | 1023 | C0-7AB |
| 36 | 1028 | DRB | 1 | . | . | 1010 | 1024 | C0-9AB |
| 37 | 1029 | DRB | 1 | . | . | 1010 | 1025 | C0-10AB |
| 38 | 1030 | DSP | 1 | . | . | 1010 | 1021 | C0-2A-3 |
| 39 | 1031 | DSP | -1 | . | . | 1010 | 1027 | C0-2AB-6 |
| 40 | 1032 | ORM | 5 | 1012 | 1017 | . | . | C1 |
| 41 | 1033 | DSP | 1 | . | . | 1032 | 1032 | C1-1 |
| 42 | 1034 | DSP | 1 | . | . | 1032 | 1032 | C1-2 |
| 43 | 1035 | DSP | 1 | . | . | 1032 | 1032 | C1-3 |
| 44 | 1036 | DSP | 1 | . | . | 1032 | 1032 | C1-4 |
| 45 | 1037 | DSP | 1 | . | . | 1032 | 1032 | C1-5 |
| 46 | 1038 | DSP | 1 | . | . | 1032 | 1032 | C1-6 |
| 47 | 1039 | DSP | 1 | . | . | 1032 | 1032 | C1-7 |
| 48 | 1040 | DSP | 1 | . | . | 1032 | 1032 | C1-8 |
| 49 | 1041 | DRB | 1 | . | . | 1032 | 1035 | C1-3A |
| 50 | 1042 | DRB | 1 | . | . | 1032 | 1037 | C1-SA |
| 51 | 1043 | DRB | 1 | . | . | 1032 | 1039 | C1-7A |
| 52 | 1044 | DRB | 1 | . | . | 1032 | 1041 | C1-3AB |
| 53 | 1045 | DRB | 1 | . | . | 1032 | 1043 | C1-AB |
| 54 | 1046 | DSP | 1 | . | . | 1032 | 1035 | C1-3A-3 |
| 55 | 1047 | DSP | 1 | . | . | 1032 | 1043 | C1-7AB-9 |
- (G)Again S1 families are screened for dwarf status and resistance and seed from single plant selection (rows 41-48 of Table 2.3.2) is kept while progeny proceed through two seasons of bulk selection (rows 49-51 for first season and rows 52 and 53 for second season in Table 2.3.2.a).
- (H) At any point, either S1 seed identified as in rows 41-48 or single plant selections from the evaluation lines, such as in rows 54 and 55 of Table 2.3.2.a can be picked by traditional breeding lines and follow normal pedigree development (rows 54 ans 55).
- (I) Subsequent ORM populations C2, C3 etc are developed and recorded exactly as for C1.
| Table 2.3.2.b: Entries in Other Progenitors Table for Open Recurrent Selection example. | |||
| Row | ID | PROGENITOR NO. | PROGENITOR ID |
| 1 | 1010 | 3 | 1006 |
| 2 | 1010 | 4 | 1007 |
| 3 | 1010 | 5 | 1008 |
| 4 | 1010 | 6 | 1009 |
| 5 | 1032 | 3 | 1019 |
| 6 | 1032 | 4 | 930 |
| 7 | 1032 | 5 | 1000 |
Genealogy Management for a Reciprocal Half-Sib Recurrent Selection Method
This section indicates how the pedigrees from the reciprocal half-sib recurrent selection method described by Fehr (1988) on page 192 can be recorded in GMS. The method is designed to simultaneously improve two populations for their combining ability with each other, hence each population is used as a broad based tester for the other. In the method described individual plants (100 or more) are identified in each population and both (1) selfed and the seed stored and (2) crossed as male to six or more individuals in the other population to form a set of half-sib families. The half-sib families are field evaluated and the individuals from each family that performed best are identified. The selfed seed from these top lines (10 or more) are grown and random mated to produce the next cycle.
- (A) In the example the pedigree starts with the formation of the 3rd cycle (Table 2.3.3.a). Reciprocal Recurrent Population A Cycle 3 (RRPAC3) is formed by population random mating (PRM) method (method 421 in Table 2.2.d) among N parents. The first two are in the GERMPLASM TABLE and the rest are in the OTHER PROGENITORS TABLE. These parents are selfed lines from the cycle 2 population A which have been identified as the top lines in the half-sib test. The GIDs of these lines will be entered not a symbolic name as here. This becomes population RRPAC3 with GID 100 (row 1 in Table 2.3.3.a). The same procedure in Population B gives RRPBC3 with GID 101 (row 2 in Table 2.3.3.a).
- (B) One hundred or more lines are identified at random (Method IDN) in each population, GIDs 201and 203 (rows 4 and 6 in Table 2.3.3.a) are examples for population A and 301 and 303 (rows 9 and 11) are examples for population B.
- (C) These lines are selfed (Method SLF) and become GIDs 202 (row 5), 204 (row 7) for population A and 302 (row 10) and 304 (row 12) for population B are examples.
- (D) Each of the lines is then mated (in this case as a male) to at least six individuals in the other population. The other population acts as a broad based tester and in this case is the female of the cross. The method is BROAD BASED TESTER, POP CF (TBP) and as the tester is the female the attribute of the method is set to reflect this and to indicate how many lines are used as females. The seed from the different females is bulked and the attributes will show how the bulking is achieved. GIDs 451 and 452 give two examples for population A (rows 14 and 15) and GIDs 551 and 552 (rows 17 and 18) for population B.
- (E) Seed from the selfed lines which are identified as superior (10 or more) are then grown in a crossing block and random mated to form cycle 4 of each population. These are indicated by GIDs 600 and 601 (rows 20 and 21 in Table 2.3.3.a).
| Table 2.3.3.a: Germplasm Table for Reciprocal Recurrent Selection example. | ||||||||
| Generative Methods | Derivative or Management Methods | Breeder’s name | ||||||
| Row in table | GID | METH | No PROGEN | FPARNT | MPARNT | GROUP | SOURCE | |
| 1 | 101 | PRM | N | RRPAC2-Sa | RRPAC2-Sb | . | . | RRPAC3 |
| 2 | 102 | PRM | M | RRPBC2-Sa | RRPBC2-Sb | . | . | RRPBC3 |
| 3 | . | |||||||
| 4 | 201 | IDN | . | . | . | 101 | 101 | RRPAC3-I1 |
| 5 | 202 | SLF | . | . | . | 101 | 201 | RRPAC3-S1 |
| 6 | 203 | IDN | . | . | . | 101 | 101 | RRPAC3-I2 |
| 7 | 204 | SLF | . | . | . | 101 | 203 | RRPAC3-S2 |
| 8 | . | |||||||
| 9 | 301 | IDN | . | . | . | 102 | 102 | RRPBC3-I1 |
| 10 | 302 | SLF | . | . | . | 102 | 301 | RRPBC3-S2 |
| 11 | 303 | IDN | . | . | . | 102 | 102 | RRPBC3-I2 |
| 12 | 304 | SLF | . | . | . | 102 | 303 | RRPBC3-S2 |
| 13 | . | |||||||
| 14 | 451 | TBP | 2 | 102 | 201 | . | . | TRRPAC3-1 |
| 15 | 452 | TBP | 2 | 102 | 202 | . | . | TRRPAC3-2 |
| 16 | . | |||||||
| 17 | 551 | TBP | 2 | 101 | 301 | . | . | TRRPBC3-1 |
| 18 | 552 | TBP | 2 | 101 | 302 | . | . | TRRPBCR-1 |
| 19 | . | |||||||
| 20 | 600 | PRM | N | 202 | 204 | . | . | RRPAC4 |
| 21 | 601 | PRM | M | 302 | 304 | . | . | RRPBC4 |
| ID = is unique germplasm identifier. METH = is the breeding method, either generative, derivative or management. No PROGEN = is the number of progenitors of a derivative method. FPARNT = ID of female parent. MPARNT = ID of male parent. GROUP = ID of last generative process for the derivative or management process. SOURCE = ID of immediate predecessor for the derivative or management process. | ||||||||
Genealogy for a polycross
The following is an illustration of how a polycross with four parents is stored in GMS. The artificial example uses clones, but inbred lines or selections from an open pollenated population could equally well have been used instead. In a polycross each line is used as a female and mated to all others and the half sib families are kept separate. In each case the other three clones are used as parents, ie. selfs are excluded. This means that the attribute for method 420 has to be set to NO. Also the other clones used as males have to be entered in the other progenitors table.
| Table 2.2.4.a: Schematic representation of a polycross with four clones as parents. | |||||
| GID | Method | Number of progenitors | Female parent | Male parent | Comments |
| 51 | 926 (cultivar release) | -1 | … | … | Clone 1 |
| 52 | 33 (unknown derivative) | -1 | … | … | Clone 2 |
| 53 | 33 | -1 | … | … | Clone 3 |
| 54 | 504 (single plant sel cf) | -1 | … | … | Clone 4 |
| 55 | 420 (polycross) | 4 | 51 | 52 | Cross with clone 1 as female. |
| 56 | 420 (polycross) | 4 | 52 | 51 | Cross with clone 2 as female. |
| 57 | 420 (polycross) | 4 | 53 | 51 | Cross with clone 3 as female. |
| 58 | 420 (polycross) | 4 | 54 | 51 | Cross with clone 4 as female. |
| 59 | 504 | -1 | 55 | 55 | Single plant selection from cross with female as clone 1. Note that group is GID = 55 and immediate source is also GID = 55. |
| 60 | 504 | -1 | 56 | 56 | Single plant selection from cross with female as clone 2. Note that group is GID = 56 and immediate source is also GID = 56. |
| Table 2.3.4.b: Other progenitors table for the polycross example. | |||
| GID | Progenitor Number | Progenitor GIID | Comments |
| 55 | 3 | 53 | Third clone used as male |
| 55 | 4 | 54 | Fourth clone used as male |
| 56 | 3 | 53 | Third clone used as male |
| 56 | 4 | 54 | Fourth clone used as male |
| … | And so for the other 2 half sib families | ||
Genealogy for a half diallel
The following is an illustration of how a half diallel with four parents is stored in GMS. The artificial example uses the same four clones as in the previous example on polycrosses, but again inbred lines or selections from an open pollenated population could equally well have been used. A half diallel is a series of single crosses with the full sib families kept separate. Each cross could equally well have been stored with the method for a single cross. The reason for recording a method for a diallel is to record extra information in the system. No attempt has been made to record every possible crossing strategy in the methods table as this would proliferate this table. Instead a compromise has been attempted between providing a minimal table (see Table 2.2.b) and information on the purposes of the matings.
| Table 2.3.5.a: Schematic germplasm table for a half diallel using the same four clones as in the example for the polycross. | |||||
| GID | Method | Number of progenitors | Female Parent | Male Parent | Comments |
| 51 | 926 (cultivar release) | -1 | … | … | Clone 1 |
| 52 | 33 (unknown derivative) | -1 | … | … | Clone 2 |
| 53 | 33 | -1 | … | … | Clone 3 |
| 54 | 504 (single plant sel cf.) | -1 | … | … | Clone 4 |
| 55 | 406 (half diallel) | 2 | 51 | 52 | Cross with clone 1 as female and clone 2 as male. |
| 56 | 406 (half diallel) | 2 | 51 | 53 | Cross with clone 1 as female and clone 3 as male. |
| 57 | 406 (half diallel) | 2 | 51 | 54 | Cross with clone 1 as female and clone 4 as male. |
| 58 | 406 (half diallel) | 2 | 52 | 54 | Cross with clone 2 as female and clone 3 as male. |
| 59 | 406 (half diallel) | 2 | 53 | 54 | Cross with clone 2 as female and clone 4 as male. |
| 60 | 406 (half diallel) | 2 | 54 | 52 | Cross with clone 3 as female and clone 4 as male. |
| 62 | 504 | -1 | 55 | 55 | Single plant selection from cross with female as clone 1 and male clone 2. Note that group is GID = 55 and immediate source is also GID = 55. |
| 63 | 504 | -1 | 56 | 56 | Single plant selection from cross with female as clone 1 and male clone 3. Note that group is GID = 56 and immediate source is also GID = 56. |
Genealogy of Rice Variety IR64
The following steps recorded in the IRRI pedigree database (Table 2.3.1.a) lead to the development of variety IR64. BCID is the breeder’s cross ID, FPARENT is the breeder’s notation for the female parent. FPSRC is the source of the female parent, either a previous BCID (defining the cross that produced the female parent, possibly by selection) or an accession number from the germplasm bank. MPARENT and MPSCR are equivalent fields for the male parent. The following illustrate many of the steps needed to enter historical genealogies.
| Table 2.3.6.a: The steps recorded in the IRRI pedigree database which led to the cultivar IR64. | ||||
| BCID | FPARENT | FPSRC | MPARENT | MPSRC |
| IR2040 | IR1561-149-1 | IR1561 | IR661-1-140-3*4/O.NIVARA | IR1737 |
| IR2055 | BPI 121-407 | 15762 | IR1416-131-5/IR576-160-2 | IR1833 |
| IR2061 | IR833-6-2-1-1 | IR833 | IR1561-149-1//IR661-1-140-3*4/O.NIVARA | IR2040 |
| IR2146 | IR773A1-36-2-1 | IR773 | IR773A1-36-2-1*3/O.NIVARA | IR1916 |
| IR5236 | IR2006-P3-31-3 | IR2006 | IR2146-68-1 | IR2146 |
| IR5338 | IR2061-465-1-4 | IR2061 | IR2055-475-2 | IR2055 |
| IR5657 | IR2006-P3-31-3/IR2146-68-1 | IR5236 | IR2061-465-1-4/IR2055-475-2 | IR5338 |
| IR18348 | IR5657-33-2-1 | IR5657 | IR2061-465-1-5-5 | IR2061 |
| IR18348-36-3-3 = IR64 | ||||
| BCID = is the breeder’s cross ID. FPARENT = is the breeder’s notation for the female parent. FPSCR = is the source of the female parent, either a previous BCID (defining the cross that produced the female parent, most likely by selection) or an accession number from the germplasm bank. MPARENT = equivalent to FPARENT for male. MPSRC = equivalent to FPSRC for male. | ||||
We can presume that the following records are already stored in the germplasm table.
| Generative Methods | Derivative or Management Methods | Breeder’s name | ||||
| ID | METH | FPARNT | MPARNT | GROUP | SOURCE | |
| 38 | C2W | 36 | 37 | . | . | IR833 |
| 68 | BC | 63 | 67 | . | . | IR1737 |
| 71 | C2W | 70 | 59 | . | . | IR1833 |
| 85 | DSP | . | . | 33 | 84 | IR773A1-36-2-1 |
| 90 | BC | 86 | 89 | . | . | IR1916 |
| 91 | ? | 81 | 60 | . | . | IR2006 |
| 93 | DSP | . | . | 57 | 92 | IR1561-149-1 |
| ID = is unique germplasm identifier. METH = is the breeding method, either generative, derivative or management. FPARNT = ID of female parent. MPARNT = ID of male parent. GROUP = ID of last generative process for the derivative or management process. SOURCE = ID of immediate predecessor for the derivative or management process. | ||||||
The new pedigrees are entered in the germplasm table as follows.
- (A) IR2040
FPARENT IR1561-149-1 is a derivative of BCID IR1561 and is already in the table with ID 93. MPARENT is a third backcross of O.NIVARA onto IR661-1-140-3 (which is incidently cultivar IR24). This backcross is IR1737, as indicated by MPSRC, and is in the table with ID 68. Hence the new cross, with ID 94, has parents 93 and 68 and BCID IR2040. From the methods table this is a single cross.
| 94 | C2W | 93 | 68 | . | . | IR2040 |
- (B) IR2055
This has a foreign female parent BPI 121-407, from a gene bank accession 15762. This is not in the germplasm table, and as nothing of its pedigree is known it can be entered as an import of an unknown source with ID 95 as follows:
| 95 | IUS | 0 | 0 | . | . | BPI 121-407 |
- The male parent is an F1 from BCID IR1833 which is in the table as with ID 71, so the new cross IR2055 can be entered as a three-way cross with immediate parents 95 and 71 as follows:
| 96 | C3W | 95 | 71 | . | . | IR2055 |
- (C) IR2061
This has female parent IR833-6-2-1-1 which is a selection out of cross IR833. The cross is in the table as ID 38, but the selection is not, so it must be added. The first step adds ID 97 for IR833-6 as a single plant selection from 38 (the first derivative in GROUP 38). The second step adds ID 98 for IR833-6-2 as a single plant selection in GROUP 38 from SOURCE 97. IDs 99 and 100 lead to IR833-6-2-1-1.
| 97 | DSP | . | . | 38 | 38 | IR833-6 |
| 98 | DSP | . | . | 38 | 97 | IR833-6-2 |
| 99 | DSP | . | . | 38 | 98 | IR833-6-2-1 |
| 100 | DSP | . | . | 38 | 99 | IR833-6-2-1-1 |
- The male parent is the F1 of the cross with BCID IR2040 which was entered as ID 94 so the process is a three-way cross. Hence we can now enter IR2061 as:
| 101 | C3W | 100 | 94 | . | . | IR2061 |
- (D) IR2146
The is the cross of a third backcross of wild species O. NIVARA onto IR773A1-36-2-1 (ID 85) which is the recurrent parent. The third backcross is identified by MPSRC as BCID IR1916 which is in the table as ID 90, so IR2146 is entered as follows:
| 102 | BC | 85 | 90 | . | . | IR2146 |
- (E) IR5236
This is a single cross between two lines derived from crosses recorded as ID 91 and ID 102. Since the derivatives are not in the table, they must be added:
| 103 | DSP | . | . | 91 | 91 | IR2006-P3 |
| 104 | DSP | . | . | 91 | 103 | IR2006-P3-31 |
| 105 | DSP | . | . | 91 | 104 | IR2006-P3-31-3 |
| 106 | DSP | . | . | 102 | 102 | IR2146-68 |
| 107 | DSP | . | . | 102 | 102 | IR2146-68-1 |
- The cross is then added as:
| 108 | C2W | 111 | 113 | . | . | IR5236 |
- (F) IR5338 is exactly similar to IR5236.
| 109 | DSP | . | . | 101 | 101 | IR2061-465 |
| 110 | DSP | . | . | 101 | 109 | IR2061-465-1 |
| 111 | DSP | . | . | 101 | 110 | IR2061-465-1-4 |
| 112 | DSP | . | . | 96 | 96 | IR2055-475 |
| 113 | DSP | . | . | 96 | 112 | IR2055-475-2 |
| 114 | C2W | 111 | 113 | . | . | IR5338 |
- (G) Next a double cross (IR5657) is made between the last two F1s:
| 115 | C4W | 108 | 114 | . | IR5657 |
- (H) IR18348
This is a single cross between derivatives from the double cross just entered (IR5657, ID 115) and the cross IR2061 ( ID 101). The derivatives of IR5657 have to be added. The male parent already has some derivatives added, IDs 109, 110 and 111. This parent is a derivative of ID 110.
| 116 | DSP | . | . | 115 | 115 | IR5657-33 |
| 117 | DSP | . | . | 115 | 116 | IR5657-33-2 |
| 118 | DSP | . | . | 115 | 117 | IR5657-33-2-1 |
| 119 | DSP | . | . | 101 | 110 | IR2061-465-1-5 |
| 120 | DSP | . | . | 101 | 119 | IR2061-465-1-5-5 |
| 121 | C2W | 118 | 120 | . | . | IR18348 |
- (I)
The last step in the development of variety IR64 consists of four generations of selection from IR18348 (ID 121):
| 122 | DSP | . | . | 121 | 121 | IR18348-36 |
| 123 | DSP | . | . | 121 | 122 | IR18348-36-3 |
| 124 | DSP | . | . | 121 | 123 | IR18348-36-3-3 = IR64 |
| Table 2.3.6.b: Germplasm Table for the genealogy of IR64. | ||||||
| Generative Methods | Derivative or Management Methods | Breeder’s name | ||||
| ID | METH | FPARNT | MPARNT | GROUP | SOURCE | |
| 38 | C2W | 36 | 37 | . | . | IR833 |
| 68 | BC | 63 | 67 | . | . | IR1737 |
| 71 | C2W | 70 | 59 | . | . | IR1833 |
| 85 | DSP | . | . | 33 | 84 | IR773A1-36-2-1 |
| 90 | BC | 86 | 89 | . | . | IR1916 |
| 91 | ? | 81 | 60 | . | . | IR2006 |
| 93 | DSP | . | . | 57 | 92 | IR1561-149-1 |
| 94 | C2W | 93 | 68 | . | . | IR2040 |
| 95 | IUS | 0 | 0 | . | . | BPI 121-407 |
| 96 | C3W | 95 | 71 | . | . | IR2055 |
| 97 | DSP | . | . | 38 | 38 | IR833-6 |
| 98 | DSP | . | . | 38 | 97 | IR833-6-2 |
| 99 | DSP | . | . | 38 | 98 | IR833-6-2-1 |
| 100 | DSP | . | . | 38 | 99 | IR833-6-2-1-1 |
| 101 | C3W | 100 | 94 | . | . | IR2061 |
| 102 | BC | 85 | 90 | . | . | IR2146 |
| 103 | DSP | . | . | 91 | 91 | IR2006-P3 |
| 104 | DSP | . | . | 91 | 103 | IR2006-P3-31 |
| 105 | DSP | . | . | 91 | 104 | IR2006-P3-31-3 |
| 106 | DSP | . | . | 102 | 102 | IR2146-68 |
| 107 | DSP | . | . | 102 | 102 | IR2146-68-1 |
| 108 | C2W | 111 | 113 | . | . | IR5236 |
| 109 | DSP | . | . | 101 | 101 | IR2061-465 |
| 110 | DSP | . | . | 101 | 109 | IR2061-465-1 |
| 111 | DSP | . | . | 101 | 110 | IR2061-465-1-4 |
| 112 | DSP | . | . | 96 | 96 | IR2055-475 |
| 113 | DSP | . | . | 96 | 112 | IR2055-475-2 |
| 114 | C2W | 111 | 113 | . | . | IR5338 |
| 115 | C4W | 108 | 114 | . | . | IR5657 |
| 116 | DSP | . | . | 115 | 115 | IR5657-33 |
| 117 | DSP | . | . | 115 | 116 | IR5657-33-2 |
| 118 | DSP | . | . | 115 | 117 | IR5657-33-2-1 |
| 119 | DSP | . | . | 101 | 110 | IR2061-465-1-5 |
| 120 | DSP | . | . | 101 | 119 | IR2061-465-1-5-5 |
| 121 | C2W | 118 | 120 | . | . | IR18348 |
| 122 | DSP | . | . | 121 | 121 | IR18348-36 |
| 123 | DSP | . | . | 121 | 122 | IR18348-36-3 |
| 124 | DSP | . | . | 121 | 123 | IR18348-36-3-3 = IR64 |
REMOTE ALLOCATION OF GERMPLASM IDENTIFIERS
The objective of ICIS GMS is to provide a tool for the management of pedigrees to a wide audience of users, many of whom do not have access to a dynamic central database. These users are often generating new entries, so the problem of the remote allocation of unique germplasm identifiers is important. Thus remote users require a copy of all GMS records up to a certain date as well as the maintenance of a local GMS, the LGMS. Each user of GMS will have a unique user ID (USER_ID) with user details stored in a user table maintained by the central ICIS administrator.
The LGMS has exactly the same structure as the GMS with internal pointers as well as pointers into the central GMS. Internal or local pointers are referred to as LOCAL IDs, TYPEs or NOs depending on the type of pointer. For example LGMS will have LOCAL_GERMPLASM_IDs, LOCAL_NAME_TYPEs, LOCAL_LOCATION_NOs, LOCAL_ATTRIBUTE_TYPEs and LOCAL_METHOD_NOs. They are only unique within the LGMS; system wide identifiers are assigned only when the central GMS is updated by each user. However the combination of USER_ID and LOCAL ID will be unique and provides the link to old records once the central GMS is updated.
The LGMS, therefore, uses LOCAL IDs, but links to progenitors or source germplasm can be IDs in the central GMS or LOCAL IDs in the LGMS. Clearly no links in the central GMS can point to LOCL IDs. LOCAL IDs must be distinguishable from central ID codes since applications must know which GMS is being used for a particular transaction otherwise the uniqueness of identifiers can become compromised through partial updates of the GMS by a user. One method of distinguishing between LOCAL IDs and central IDs would be to make all the former ones negative.
Users would periodically send subsets of LGMS to the GMS administrator for update. These subsets could be defined as all records up to a specified date. The GMS administrator would add the LGMS records for all tables to GMS assigning new ID codes, and adding USER_ID and LOCAL_GERMPLASM_ID to germplasm records so that entries in other private databases such as the SGM or LDMS (local data management system) are automatically linked to GMS through a search on these fields and do not need to be explicitly updated. This implies that USER_IDs and LOCAL IDs must never be reused even after updating. After an update the user gets a new version of GMS, and the LGMS is scrubbed of all updated records. Thereafter, any search of LGMS for a LOCAL_GERMPLASM_ID which has already been updated triggers a search in the updated GMS for USER_ID and LOCAL_ID.
UPDATING THE CENTRAL GMS
Presumably local users could update as soon as set generation is complete. Would Set Records then be updated before going to FBM? Clearly this cannot be a requirement because we must cater for remote users. What about users of the central ICIS DMS? Do they have to update before data entry? Presumably yes. Then any LOCAL ID encountered is known to be an attribute and its ID can be found by an attribute search at data entry. Most remote users will probably not send data when they update GMS.
Is it necessary to keep Set Records? Yes probably but not centrally, because if a user does not use ICIS FBM and DMS then set records are the only ICIS format data available for specifying progenitors or selections by choosing from previous trials. If they are kept, is it necessary to update them when GMS is updated? Probably not because any lists dated prior to the last update of the GMS are known to contain LIDs as attributes, while later ones still contain ID pointers.
REFERENCES
Fabrizius, M. A., Cooper, M. Podlich, D., Brennan, P.S., Ellison, F.W. and DeLacy, I.H. 1996. Design and simulation of a recurrent selection program to improve yield and protein in spring wheat. University of Queensland, Dept. of Agriculture, Brisbane, QLD 4072 Australia.
Fehr, W. R. 1987. Principles of cultivar development. Vol 1. Theory and Technique. Macmillan Publishing Company, New York.