TDM Genealogy Management System
From ICISWiki
PHYSICAL STRUCTURE OF THE ICIS GMS DATABASE
INTRODUCTION
The ICIS GMS database stores and manages information on genesis, genealogy, nomenclature and chronology of germplasm for a particular crop. It operates on the principle of unique identification of germplasm through system assigned Germplasm IDs (GIDs). It also manages the ambiguities of synonyms and homonyms through name searching facilities, and it allows remote allocation of unique GIDs by managing a user list and operating through a twin database system comprising a copy of the Central GMS and a Local GMS with identical structure. Any DBMS program, which supports Open Database Connectivity (ODBC), can be used to run GMS, and each implementation of local GMS can use a different ODBC compliant DBMS. The DBMS for the central GMS can be chosen independently for each crop, and read-only distribution copies for access by local installations need not use the same DBMS as the actual central GMS. All user access to the databases is through a suite of access routines provided in the form of a Windows 95 32-bit Dynamic Link Library (DLL). Basic applications for querying, updating and reporting will be written in the most convenient language for the application, and users will be able to write their own applications by calling the DLL. Any such programs will operate identically on any GMS installation of any ICIS implementation.
The Central GMS
The ICIS administrator for a particular crop sets up the central GMS, it comprises sixteen tables: four data tables indexed by GIDs, three look-up tables to manage references, methods constant codes and user defined fields, three tables to manage users, installations and structural changes, and six to manage locations. Only the administrator has write access to the central GMS, users can read it, but all user updates are recorded on an installation of local GMS and must be passed through the administrator to be entered on the central GMS and hence be accessible to other users.
The Local GMS
Each installation of GMS has a number of local users with read-only access to central GMS, or to a copy of it, and write access to a local GMS. The local administrator has complete write access to every table of local GMS and other users have different levels of access assigned by the local administrator (3.8.3). The local GMS has the same structure as central GMS except that three implementation specific tables are not included and list management tables are added (3.10).
Discrimination between central GMS and local GMS
The GMS user will be completely unaware that there are two databases operating. However the internal system must be able to distinguish between the two, and this is achieved by the local GMS always using negative key fields, and the central GMS always using positive key fields. Links to these key fields from any tables in local GMS can be positive or negative depending on whether they refer to a record in the central GMS or the local GMS. Links in the central GMS are always positive since the central GMS is 'unaware' of records in individual installations until they have been updated.
Allocation of USER IDs
The ICIS central administrator will allocate a set of unique USER IDs for each installation. This ensures the uniqueness of remotely assigned Germplasm IDs. Each installation of GMS will have a local administrator who can assign the USER IDs to specific users by specifying information in the USERS table (3.8.2). Once filled, the central administrator can only change these fields. All users of a local installation will have read access to all records of the local GMS.
Data Types
The only data types used in GMS are long and short signed integers and character strings except for latitude (LAT), longitude (LON) and altitude (ALT) in the GEOREF table (3.7.6) which are eight byte or 4 byte (ALT) floating point real fields. Dates are stored as long integer (4 bytes) as follows: YYYYMMDD. If the day or month and day are unknown then DD or MM are set to 0. If day is known neither month nor year should be missing. Also if month is known then year should not be missing. User defined data structures always store ASCII text but users may specify a format to convert these strings to data structures with elements of integer, or text types (3.6.3). Specialist application programs can then use this data as required. Time can be stored in user defined data structures and should be stored as HHMMSS in twenty-four hour format.
Missing Values
Valid values must be provided for all fields unless the admissibility of null values is specifically described in the SCHEMA. In some cases, valid data for fields includes values like 'UNKNOWN', which are not missing. Missing values referred to in the SCHEMA depending on the data type of the field. Missing values for integer fields, when permissible, are zero. Missing values for text fields are null strings and missing values for float fields are -1.0E36.
User Definition of Constants and Data Fields
The database structure allows users to define new methods, data fields and add data types at any time through a system of field definitions. This means that new methodology in germplasm development can be accommodated, and new types of information can be stored when required.
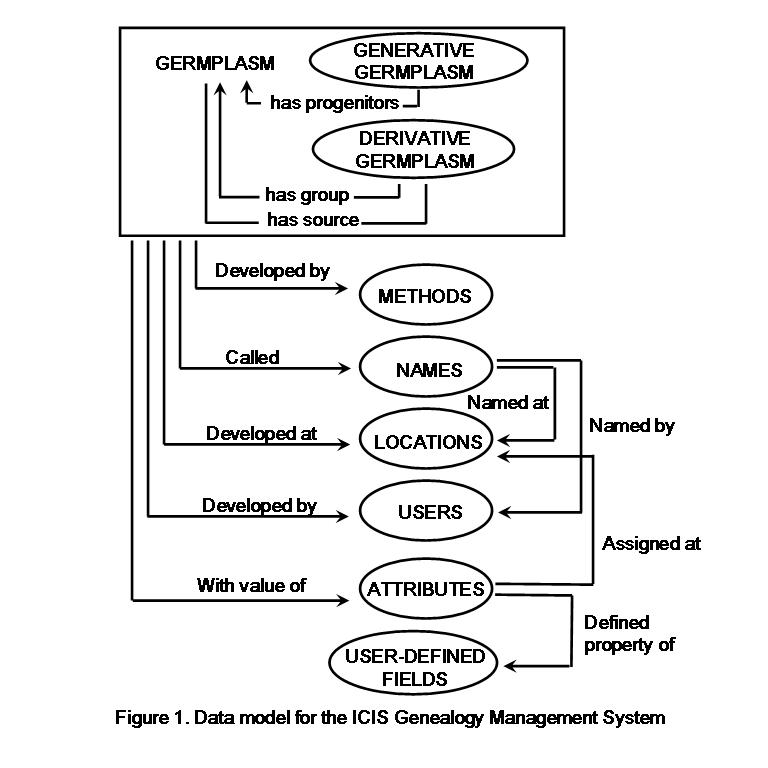
GENEALOGY MANAGEMENT SYSTEM
GERMPLASM TABLE (GERMPLSM)
Main details of germplasm genesis, origin and preferred name
| Columns - Long Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| GERMPLSM_ID (GID) | Unique germplasm identifier. GID is automatically generated by the system each time a new record is added. Links to GID in PROGNTRS, NAMES and ATRIBUTS tables | Number (Long) | 4 |
| NO_PROGENITORS (GNPGS) | Defines type of genesis and number of progenitors.
For a derivative or maintenance process GNPGS =-1 and then GPID1 contains the germplasm group ID and GPID2 the source germplasm ID. For a generative process GNPGS contains the number of specified parents. (The number of parents required by a method is recorded by NPRGN on the METHODS TABLE). If GNPGS=1 or 2 then the IDs of the progenitors are contained in the GPID1 and GPID2 fields on the GERMPLSM table. If GNPGS>2 then further IDs are stored on the PROGNTRS table. GNPGS = 0 for land race or wild species collections or if none of the parents is known. GNPGS <= NPRGN, but some of the GNPGS specified parents may be unknown in which case the corresponding GPIDs are MISSING (0). For example in a simple cross with only male parent known, GNPGS would have to be 2 with GPID1=0 and GPD2 set to GID of the known male parent. | Number (Integer) | 2 |
| METHOD_NO (METHN) | Number that identifies the method of genesis for the germplasm. Details of the method are on the METHODS table and specific parameters may be stored on the ATTRIBUTES table (see MATTR on the METHODS table). Links to MID in METHODS table. | Number (Integer). | 2 |
| PROGENITOR_ID1(GPID1) | If GNPGS is -1 then GPID1 is the group ID, which is the GID of last source germplasm produced by a generative process, or the GID of the last source of unknown origin in the development path of the current germplasm. This ID defines a group of germplasm in which all members are derived from the same generic parent.
If GNPGS >0 then GPID1 points to the first progenitor in a generative process (usually the female parent). GPID1=0 if unknown. | Number (Long) | 4 |
| PROGENITOR_ID2 (GPID2) | If GNPGS is -1 then GPID2 points to the immediate source from which the current germplasm is derived
If GNPGS>0 then GPID2 points to the second progenitor of the germplasm in a generative process (usually the male parent). GPID2=0 if unknown. | Number (Long) | 4 |
| MANAGEMENT_GID (MGID) | If the current germplasm is a managed sample then MGID contains the GID of the germplasm at the root of the management tree, else 0. | Number (Long) | 4 |
| GERMPLASM_USER_ID (GERMUID) | ID of the user entering details of the current germplasm. UID in the USERS table | Number (Integer) | 2 |
| LOCAL_GERMPLASM_ID (LGID) | LGID contains the original, negative GID used in the Local GMS prior to transfer of data records to the central GMS. LGID = 0 if the record was not entered through an update process | Number (Long) | |
| LOCATION_NO (GLOCN) | Identifier of the location where the germplasm was created as a distinct unit of management with a new GID. LOCID in LOCATION table. GLOCN = MISSING or 0 if location is unknown
N.B. GIDs derived only by maintenance methods from a sample collected from a farm, market place etc, should have their GPID1 point to the GID of the original collected sample, and GLOCN of the collected sample is the collecting location. | Number (Long) | 4 |
| GERMPLASM_DATE (GDATE) | Date of germplasm genesis, i.e. the date on which the germplasm was created as a distinct unit of management with a new GID. (YYYYMMDD). GDATE = 0 if unknown.
| Number (Long) | 4 |
| GERMPLASM_REFERENCE (GREF) | A number that identifies a bibliographic reference from where the germplasm data was retrieved. GREF is missing if the source is unpublished or unknown | Number (Long) | 4 |
| GERMPLASM_REPLACE (GRPLCE) | Records deletion or replacement for the current GERMPLASM record. 0 for unchanged, own GID for deleted, and replacement GID for replaced | Number (Integer) | 4 |
OTHER_PROGENITORS TABLE (PROGNTRS)
Stores IDs of progenitors for generative methods that require more than two parents
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| GERMPLASM_ID (GID) | Identifier for the germplasm. GID in the GERMPLSM table | Number (Long) | 4 |
| PROGENITOR_NO (PNO) | Progenitor number 3, 4, ... | Number (Integer) | 2 |
| PROGENITOR_ID (PID) | Germplasm identifier for progenitor number PNO. GID in the GERMPLSM table. PID = 0 if the progenitor is unknown | Number (Long) | 4 |
DEFINITION OF METHODS
Method definitions are stored in the METHODS table, and complete documentation, including bibliographic references, is expected. One anticipated use of the methods table is to provide information to applications which calculate the degree of relationship between germplasm, or which trace gene flows through germplasm development. Some methods depend on parameters, which may vary each time the method is used, such as the number and mixing proportions of parents in the generation of a population. These parameters can be defined by assigning an attribute to the method with the METHOD_ATTRIBUTE field. Values for each instance of the associated method are recorded on an attribute record linked to the GID of the germplasm being produced. Attributes are flexible user definable data fields (3.5).
The METHODS TABLE (METHODS)
Details of methods of germplasm genesis
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| METHOD_ID (MID) | Identifier for the method of germplasm development. METHN on the GERMPLSM table. | Number (Integer) | 2 |
| METHOD_TYPE (MTYPE) | Method type: GEN for generative, DER for derivative, MAN for maintenance methods | Text | 3 |
| METHOD_GROUP (MGRP) | Defines the breeding system to which the method applies: S – self fertilizing, O – cross pollinating, C clonally propagating and G – all systems | Text | 3 |
| METHOD_CODE (MCODE) | Mnemonic description of method | Text | 8 |
| METHOD_NAME (MNAME) | Name of method | Text | 50 |
| METHOD_REFERENCE (MREF) | Number of a reference to the method. Links to the Reference number in the Bibliography Table. May be missing | Number (Long) | 4 |
| METHOD_DESCRIPTION (MDESC) | Description of the method. | Text | 255 |
| NO_PROGENITORS (MPRGN) | MPRGN =-1 if the method is derivative or management. Otherwise MPRGN is the number of progenitors required for the generative process. MISSING (0) if the number of progenitors is variable | Number (Integer) | 2 |
| NO_FEMALE_PARENTS (MFPRG) | Number of female parents for a generative process, these are specified first in the list of parents. MPRGN = MISSING if the method is derivative or the number of female parents is variable (if variable it can be recorded as an attribute for each instance, see MATTR). | Number (Integer) | 2 |
| METHOD_ATTRIBUTE (MATTR) | Identification number of an attribute associated with this method. The attribute is defined in the UDFLDS table. FLDNO in UDFLDS table and ATYPE in ATRIBUTS table. MISSING if no attribute is associated with the current method. | Number (Integer) | 2 |
| GENETIC_EQUIVALENCE (GENEQ) | METHN of a basic method which has equivalent genetic relationship between progenitors and offspring for the purpose of computing coefficients of parentage | Number(Integer) | 2 |
| METHOD_USER (MUID) | ID of the user defining the method | Number (Integer) | 2 |
| LOCAL_METHOD_ID (LMID) | Local method number from the installation where it was defined | Number (Integer) | 2 |
| METHOD_DATE (MDATE) | Date of method definition | Number (Long) | 4 |
NAME DATA
Germplasm collects a multitude of labels during the development and release process. These are all tracked through the NAMES table of GMS. Names are classified by name types which are defined in the USER_DEFINED_FIELDS table (3.6). An abbreviation may be associated with each name type, they have a maximum length of eight characters and are stored in the NAME field of the NAMES table like any other name. They are defined in the USER_DEFINED_FIELDS table with USER_FIELD_TYPE = ABBREVIATION but with the same name type as the name Abbreviations inherit the NAME_TYPE, NAME_USER, NAME_LOCATION and NAME_REFERENCE values from the name being abbreviated. This allows abbreviations to be matched to names. Unique matching requires the restriction that different names cannot have all these fields identical. Abbreviations take on their own NAME_DATE so that multiple abbreviations of the same name can be distinguished. One name must be defined as the preferred name, one abbreviation may also be declared as preferred (this need not be an abbreviation of the preferred name). Names may contain imbedded information, and this can be made accessible to application programs for specific name types by specifying a format for the imbedded information in the USER_DEFINED_FIELDS table (3.6.3). All names for local germplasm must appear in the local names table, but some names for germplasm in the central database can appear in the local names table (with positive GID values). One name in the central must be preferred, but central germplasm may also have one name set as preferred in the local names table. This name takes precedence over the preferred name in the central database whenever the particular local is being used. This allows local users to change preferred names or specify local preferred names.
The Names Table (NAMES)
Stores all germplasm names, abbreviations and naming details.
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| NAME_ID (NID) | Unique identifier for the name | Number (Long) | 4 |
| GERMPLASM_ID (GID) | Identifier for the germplasm associated with the name. GID in GERMPLSM table | Number (Long) | 4 |
| NAME_TYPE (NTYPE) | Number that identifies the type of name or abbreviation. FLDNO in UDFLDS table | Number (Integer) | 2 |
| NAME_STATUS(NSTAT) | Number indicating the storage type and status of the name: 1 – Preferred name (must be ASCII), 8 - Preferred ID, 2 - Preferred abbreviations (must be ASCII), 8- Preferred ID, 3 - Chinese-GBK (GD) DBCS names, 4 - Chinese Big 5, 5 - Japanese 6 – Korean, 10 - UNICODE names which are not preferred, 9 - the name is marked as deleted | Number (Integer) | 2 |
| NAME_USER_ID (NUID) | ID of the user naming the germplasm. UID in the USERS table | Number (Integer) | 2 |
| NAME_VALUE (NVAL) | Germplasm name.
For ASCII names each character occupies one byte, but for UNICODE names (eg Chinese), each character occupies 2 bytes | Text | 128 |
| NAME_LOCATION_NO (NLOCN) | Identifier for the location where the name was first assigned to the maintenance neighbourhood of which this GID is a member. LOCN in LOCATION table. NLOCN = 0 if the location is unknown.
| Number (Long) | 4 |
| NAME_DATE (NDATE) | Date on which the name was first assigned to the maintenance neighbourhood of which this GID is a member. (YYYYMMDD). NDATE = 0 if the date is unknown
| Number (Long) | 4 |
| NAME_REFERENCE(NREF) | A number that identifies a bibliographic reference for the name. NREF is missing if the source is unpublished or unknown | Number (Long) | 4 |
Name Standardization
A major problem with identifying germplasm is the detection of minor variants of a name. For example some users hyphenate foreign language names while others use spaces or capital letters within a name string of small case letters, some separate a character prefix from a number by a space, others do not. Another problem is that different ODBC drivers implement the same SQL search differently with respect to case sensitivity. For these reasons, a set of name standardization has been devised with the objective of producing the same standardized name from as wide a range of variants as possible. These rules are applied in order as follows:
(L= any letter; ^= space; N= any numeral, S= any of {-,',[,],+,.})
a) Capitalize all letters Khao-Dawk-Mali105 becomes
KHAO-DAWK-MALI105
b) L( becomes L^( and )L becomes )^L IR64(BPH) becomes IR64 (BPH)
c) N( becomes N^( and )N becomes )^N IR64(5A) becomes IR64 (5A)
d) L. becomes L^ IR 63 SEL. becomes IR 64 SEL
e) LN becomes L^N EXCEPT SLN MALI105 becomes MALI 105
but MALI-F4 IS unchanged
f) NL becomes N^L EXCEPT SNL B 533A-1 becomes B 533 A-1
but B 533 A-4B is unchanged
g) LL-LL becomes LL^LL KHAO-DAWK-MALI 105 becomes
KHAO DAWK MALI 105
h) ^0N becomes ^N IRTP 00123 becomes IRTP 123
i) ^^ becomes ^
j) REMOVE LEADING OR TRAILING ^
k) ^) becomes ) and (^ becomes (
l) L-N becomes L^N when there is only one ‘–‘ in the name and L is not preceded by a space
m) ^/ becomes / and /^ becomes /
When a name string is supplied to the ICIS DLL for searching, it is searched as supplied and after standardization. Routines, which write names to the database, do not apply the standardization and users must specifically call a standardization routine if they wish to standardize names before storing them. Names in the database are not required to be standardized. It is hoped that the resulting standardized name is acceptable for presentation, but there will be cases where users insist on having a name, which violates the rules. For example CIMMYT prefers that abbreviations contain no spaces. This is allowed but the rule violation should not be used to distinguish genotypes, and the standardized name should be given as a synonym. Users will always be able to search for names that violate the rules and the correct germplasm should be found.
GERMPLASM ATTRIBUTES
Attributes are text variables used to store information about the genesis, genealogy, nomenclature or chronology of germplasm. Attribute types are defined and described on the USER_DEFINED_FIELDS table (3.6.1). Like names, attributes may contain imbedded information in the form of sub-fields or variables within the attribute text. The structure and format of such attributes is defined through the USER_FIELD_FMT of the USER_DEFINED_FIELDS table, as is the format of structured names (3.6.3).
The ATTRIBUTES TABLE (ATRIBUTS)
Table of germplasm attributes used to store information about germplasm genesis, genealogy and nomenclature, which is not stored in other tables.
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| ATTRIBUTE_ID (AID) | ID of the Attribute (unique) | Number (Long) | 4 |
| GERMPLASM _ID (GID) | ID of germplasm with which the attribute is associated. GID in GERMPLASM table. | Number (Long) | 4 |
| ATTRIBUTE_TYPE (ATYPE) | Number that identifies the attribute. FLDNO in UDFLDS table. Use 999 to mark the attribute as deleted. | Number (Integer). | 2 |
| ATTRIBUTE_USER_ID (AUID) | ID of the user giving the attribute value. UID in USERS table | Number (Integer) | 2 |
| ATTRIBUTE _VALUE (AVAL) | Contains the attribute value. | Text | 255 |
| ATTRIBUTE_LOCATION_NO (ALOCN) | Identifier for the location where the attribute value was set. LOCN on the LOCATION table. ALOCN = MISSING if the location is unknown. | Number (Long) | 4 |
| ATTRIBUTE_REFERENCE(AREF) | Key to the reference for the attribute data. MISSING if the source is unknown | Number (Long) | 4 |
| ATTRIBUTE_DATE (ADATE) | Date of value assignation. (YYYYMMDD) | Number (Long) | 4 |
USER DEFINED CONSTANTS AND DATA FIELDS
The ICIS structure allows considerable flexibility in the type and scope of information that can be stored. It does this by storing structural information (metadata) in the USER DEFINED FIELDS TABLE (UDFLDS). Some of this structural information is standard across implementations and is pre-loaded, some is specific to particular crops and is loaded by the central database administrator, and some is specific to local installations and can be loaded or modified by individual users. Types of metadata range from the definition of constants, where only the range or scope of allowable values is defined, to definition of fields where the type, scope and structure are defined. Allowable user or reference types are examples of constants and attribute or location descriptors are fields since they include both description and structure. In some cases both types are possible. For example name types may also have a format describing the structure of information imbedded in a name of a particular type.
The USER_DEFINED_FIELDS TABLE (UDFLDS)
Table of definitions of user defined fields and constants
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| USER_FIELD_NO (FLDNO) | Unique identifier for the user defined field. NTYPE in NAMES table, ATYPE in the ATRIBUTS table and MATTR in the METHODS table, DTYPE in the LOCDES table and to the table and field specified in FTABLE and FTYPE for constant definitions | Number (Integer) | 2 |
| USER_FIELD_TABLE (FTABLE) | Short Name of the ICIS data table to contain the user defined field:
NAMES, ATRIBUTS or LOCDES or the table containing the constant link | Text | 24 |
| USER_FIELD_TYPE (FTYPE) | Type of defined field: NAME, ATTRIBUTE, ABBREVIATION, or DESCRIPTOR or field name for definition of constants | Text | 12 |
| USER_FIELD_CODE (FCODE) | Short name for the user defined field or a permissible values for a constant | Text | 8 |
| USER_FIELD_NAME (FNAME) | Long name for the user defined field or descriptive value for a constant | Text | 50 |
| USER_FIELD_FMT (FFMT) | Definition of sub-fields or variables contained in the user defined field. These variables can have multiple elements, and may contain text, integer or real valued data. (See section 3.6.3). If FFMT is blank then the user-defined field contains a single, unnamed text variable with variable length such as a name. | Text | 255 |
| USER_FIELD_DESCRIPTION (FDESC) | Full description of the user defined field, which must describe all sub-fields specified in FFMT and may contain a bibliographic reference. | Text | 255 |
| LOCAL_FIELD_NO(LFLDNO) | Local field number | Integer | 2 |
| FIELD_USER_ID(FUID) | ID of the user defining the field | Integer | 2 |
| FIELD_DATE(FDATE) | Date of field definition | Integer | 4 |
CONSTANT Values
Many fields in ICIS tables are restricted to a discrete set of possible values. The values actually stored in the tables may be field-Ids (FLDNO) or field-codes (FCODE) from the USER_DEFINED_FIELDS table. The full set of possible values for such a field is indexed by FTABLE, indicating the table where they are stored, FTYPE, the name of the field in the table, and FCODE, the actual values of the constant. The FNAME field contains a longer description of the constant value. Users can extend the range of possible values allowed for a constant by adding records to the local USER_DEFINED FIELDS table. An example is the set of location types that are recorded in the TYPE field of the LOCATION table. The associated constants in the UDFLDS table are:
CONSTANT Field Values
| Columns - FLDNO | FTABLE | FTYPE | FCODE | FNAME |
|---|---|---|---|---|
| 401 | LOCATION | LTYPE | 1 | CONTINENT |
| 402 | LOCATION | LTYPE | 2 | GEOGRAPHICAL REGION |
| 403 | LOCATION | LTYPE | 3 | GEOPOLITICAL REGION |
| 404 | LOCATION | LTYPE | 4 | ECOLOGICAL REGION |
| 405 | LOCATION | LTYPE | 5 | COUNTRY |
| 406 | LOCATION | LTYPE | 6 | FIRST SUB-NATIONAL DIVISION |
| 407 | LOCATION | LTYPE | 7 | SECOND SUB-NATIONAL DIVISION |
| 408 | LOCATION | LTYPE | 8 | THIRD SUB-NATIONAL DIVISION |
| 409 | LOCATION | LTYPE | 9 | GERMPLASM COLLECTION SITE |
| 410 | LOCATION | LTYPE | 10 | BREEDING LOCATION |
| 1401 | LOCATION | LTYPE | INGER | INGER TEST SITE |
The first ten constant values are standard across implementations, the last is specific to IRIS and hence its ID is greater that 1000. Further constant values can be added to local UDFLDS tables with negative Ids.
Name and Attribute Descriptor Formats
Sub-fields or variables contained within user defined names or attributes are specified with the USER_FIELD_FMT (FFMT) field of the USER_DEFINED_FIELDS table. Variables may consist of different data types, and have multiple elements as in arrays. The values of all variables in names or attributes are stored in ASCII representation in the order they are defined. The format specifies how the variables may be extracted from the text string. Each user-defined variable is described in consecutive statements of FFMT in the order that the values will be stored. Each statement consists of four items separated by commas. Semicolons separate statements. Each variable definition appears as follows: NAME, N, T, COUNT; where:
- NAME contains a name for the variable,
- N defines the text length of each element of the variable,
- T defines the data type and
- COUNT defines the number of elements in the variable.
- NAME is an alphanumeric value with first character alphabetic. It can be blank or absent to indicate unnamed space filler.
- N gives the number of characters occupied by each element of the variable if it is an integer value. N can also consist of a character contained in double quotes defining the delimiter between elements. If the last variable is a single element text variable, then N may be zero, blank or absent to indicate variable length for that variable.
- T can have value A for ASCII text data, I for integer data and R for real valued data, S for an integer sequence or a string of length N in quotes for a constant. IF T is blank or absent A is assumed.
- COUNT may be a positive integer defining the number of elements or it may be the name of a previously defined, single-element, integer variable whose value will define the number of elements or an integer of N digits defining the next sequence number if T has the value S. If COUNT is blank then the variable is assumed to have a single element.
Example 1
A selection method may record the number and the names of the stresses under which selection was practised, and a breeder’s comment regarding the selection.
The format record would look as follows:
- NSTRESS,4,I,1;STRESS,12,A,NSTRESS; COMMENT,0,A,1
or
- NSTRESS,4,I;STRESS,12,A,NSTRESS; COMMENT
or
- NSTRESS,4,I;STRESS,”,”,A,NSTRESS;COMMENT
Example 2
A germplasm name-type can be defined to contain sub-fields. These sub-fields are ignored during all GMS name manipulation but may be useful to specific application programs. As an example, a name could contain a two-letter program ID, a year code, a five character cross designation and a selection string as follows: UR96IR123 10 3W 4. The format line would look as follows:
- PROGRAM,2,A,1;YEAR,2,I,1;CROSS,5,A,1;SELN,0,A,1
or
- PROGRAM,2;YEAR,2,I;CROSS,5;SELN
Example 3
A name type could have a constant Prefix, a sequence number and a suffix. IF the next number in the sequence is 10001 then the FFMT field would be:
- PREFIX,3,’CLT’,1;NEXT,5,S,10001;SUFFIX,1,’H’,1
The next name assigned with this type would be CLT10001H and the FFMT statement in the UDFLDS table would be updated to PREFIX,3,’CLT’,1;NEXT,5,S,10002;SUFFIX,1,’H’,1.
CORRECTING RECORDS IN THE GMS DATABASE
Corrections and changes will inevitably occur in any database. Authorised users can only make these changes, and all changes are logged so that the sequence of changes can be traced and can be ‘undone’ if required. The history of changes is tracked by records on the CHANGES table. One common occurrence of changes is when new information about an existing germplasm record is entered into a local database. If the existing record is also in the local database then the local user can complete the changes. But if it is in the central database, then changes cannot be completed until the central database is updated. Verifying and completing requested changes is part of the update process and sufficient information and justification needs to be recorded in the changes table to allow the process to be completed. Verification and completion of changes to the central database may take some time but local users would like to see their changes reflected immediately. This is achieved by the DLL which always checks the local CHANGES table for central changes and applies them at run time for the specific installation where they are recorded.
The CHANGES Table
| Columns - Long Name (Name) | Description | Type | Length (bytes) |
|---|---|---|---|
| CHANGE ID (CID) | Unique change identification number | Integer | 4 |
| CHANGE_TABLE (CTABLE) | Name of the table where the change is to be effected | Text | 16 |
| CHANGE_FIELD (CFIELD) | Name of the field where the change is to be effected | Text | 16 |
| CHANGE_RECORD (CRECORD) | Key ID of the record where the change is to be effected | Integer | 4 |
| CHANGE_FROM (CFROM) | Previous value of the field | Integer | 4 |
| CHANGE_TO (CTO) | New value of the field | Integer | 4 |
| CHANGE_DATE (CDATE) | Date of the change (YYYYMMDD) | Integer | 4 |
| CHANGE_TIME (CTIME) | Time of the change (HHMMSSMM) | Integer | 4 |
| CHANGE_GROUP | Group identification for dependant changes(Eg DATE/TIME string) | Text | 20 |
| CHANGE_USER (CUID) | Id of the USER making the change | Integer | 2 |
| CHANGE_REFERENCE (CREF) | Chang reference | Integer | 4 |
| CHANGE_STATUS | Change status: 0=ACTIVE, 1=UNDONE, 2=UPDATED | Integer | 4 |
| CHANGE_DESCRIPTION (CDES) | Change description or reason | Text | 255 |
Change location, date or reference of germplasm
To change location, date or reference of germplasm with GERMPLASM_ID GIDX.
Checks:
a) Check that GIDX has not been deleted or replaced.
Changes:
a) Add a change record to the local database with the following values:
CID - Next available negative integer
CTABLE - GERMPLSM
CFIELD - GLOCN, GDATE or GREF as appropriate
CRECORD - GIDX
CFROM - Current value of the field
CTO - New value for the field
CDATE - Current date
CTIME - Current time
CGROUP - Null
CUID - Current user ID.
CSTATUS - 0
CDESC - Reason for change
b) If the germplasm is local change the required field.
c) (a)-(b) can be repeated for more than one field.
Change a progenitor ID for generative germplasm
Change the ID of progenitor number N of germplasm with GERMPLASM_ID GIDX from GIDY to GIDZ (GIDZ may be MISSING (0) to indicate that progenitor number N is unknown)
Checks:
a) Check that germplasm GIDX is produced by a generative process and has at least
N progenitors (NO_PROGENITORS ≥ N) b) Check that GIDZ precedes GIDX if it is not MISSING.
Changes:
a) Add a record to the CHANGES table.
IF N is 1 or 2, the first five field of the CHANGE record will have values:
CID: Next available negative integer
CTABLE: GERMPLSM
CFIELD: GPID1 or GPID2 depending on whether N is 1 or 2
CRECORD: GIDX
CFROM: GIDY
CTO: GIDZ
If N is > 2 the the fields will have the following values:
CID: Next available negative integer
CTABLE: PROGNTRS
CFIELD: PIDx where x=N
CRECORD: GIDX
CFROM: GIDY
CTO: GIDZ
b) If GIDX < 0 change PROGENITOR_ID for progenitor number N on the GERMPLASM table
or the OTHER_PROGENITORS table from GIDY to GIDZ.
c) If GIDZ is missing and N=NO_PROGENITORS then reduce NO_PROGENITORS by 1 and
if N>2 delete the record from the OTHER_PROGENITORS table.
Add a progenitor ID for a generative germplasm
Add the germplasm GIDZ as a PROGENITOR_ID for germplasm GIDX.
Checks:
a) Check that germplasm GIDX is produced by a generative process and determine
the number, N, of progenitors specified, N=NO_PROGENITORS. b) Check that the generative method of GIDX supports N+1 progenitors. c) Check that GIDZ precedes GIDX
Changes:
a) Add a CHANGE record with CTABLE=GERMPLSM, CFIELD=GNPGS, CRECORD=GIDX, CFROM=N,
CTO=N+1. Then add a second CHANGE record as in 3.9.3.a with GIDY=0. b) If GIDX<0 change NO_PROGENITORS to N+1 in the local GERMPLSM table. c) If GIDX<0 and N+1 is 1 or 2, change PROGENITOR_ID1 or PROGENITOR_ID2 from MISSING
to GIDZ. IF N+1 > 2 add a record to the OTHER_PROGENITORS table
the GERMPLASM_ID=GIDX, PROGENITOR_NO=N+1 and PROGENITOR_ID=GIDZ.
Change the group ID for a derivative germplasm
To change the group ID of GIDX from GIDY to GIDZ take the following steps:
Checks:
a) Check the germplasm GIDX is derivative. b) Check that GIDZ is generative or a derivative with unknown source (GPID1
and GPID2=0) c) Check that the new group precedes the target in chronology. d) If GIDZ # 0, the new group of GIDX (GGPX) will be GIDZ. Check the source of
GIDX has group GIDZ. If not, set the source to zero (3.9.6) or change its group
to GIDZ using this algorithm. e) If GIDZ = 0, then GIDX will become a derivative with unknown source (GPID1=0)
so its group, GGPX, is GIDX. Check that the source (GPID2) is also zero. f) Check that all dependants of GIDX have group GIDZ.
Changes:
a) Add a CHANGE record with CTABLE=GERMPLSM, CFIELD=GPID1, CRECORD=GIDX CFROM=GIDY,
CTO=GIDZ and put a unique group identifier
such as the DATE/TIME string in CGROUP. b) If GIDX<0 change PROGENITOR_ID1 for germplasm GIDX on the GERMPLSM table
from GIDY to GIDZ. c) Check all derivatives with derivative paths tracing through GIDX and change
their groups to the new group of GIDX, GGPX, using this algorithm. These dependant
changes get the same CGROUP string as the original change.
Change a source ID for a derivative germplasm
To change the ID of the source for germplasm GIDX from GIDY to GIDZ
Checks:
a) Check that a derivative process produces GIDX. b) Check that GIDZ precedes GIDX. c) Check that GIDZ is from the same germplasm group as GIDX. If it is not,
then the group IDs of all dependants will have to be traced and changed. d) Check that the selection name for GIDX does not indicate a source other than GIDZ.
Changes:
a) Add a CHANGE record with CTABLE=GERMPLSM, CFIELD=GPID2, CRECORD=GIDX,
CFROM=GIDY, CTO=GIDZ and CGROUP=date/time. Other Change fields have appropriate values. b) If GIDX<0 change PROGENITOR_ID2 for germplasm GIDX on the local GERMPLASM
table from GIDY to GIDZ. c) If GIDZ # 0, check the group of GIDX is the same as the group of GIDZ.
If not, change the group of GIDX to the group ID of GIDZ (3.9.5) and check
the group ID of all dependants of GIDX (direct derivatives and derivatives in the same
group with development paths tracing through GIDX). d) If GIDZ = 0, and the group of GIDX # 0 then check whether the
group of GIDX should also be set to 0 (missing).
Change a germplasm method
To change the method of germplasm GIDX from METH1 to METH2:
Checks:
a) Check that the new method supports the number of progenitors of GIDX b) Check that the progenitors are appropriate for the new method of GIDX.
Changes:
a) Add a CHANGE record with CTABLE=GERMPLSM, CFIELD=METHN, CRECORD=GIDX,
CFROM=METH1, CTO=METH2, CGROUP=date/time string and other fields have
appropriate values. b) If the method is being changed from derivative to generative then
the group IDs of all derivatives with paths tracing through GIDX need to change
to GIDX (3.9.5). c) If the method is being changed from generative to derivative all
derivatives in the group GIDX need to change group ID to the new group of GIDX.
(This may be GIDX if it becomes a derivative group source) (3.9.5). d) If the number of progenitors change or the method type changes
add a CHANGE record with CTABLE=GERMPLSM, CFIELD=GNPGS, CRECORD=GIDX, CFROM=old value,
CTO=new value, CGROUP=same string as in a. e) If GIDX<0 make the changes in the local germplasm table
Replace an existing germplasm record with another one
This change is required when germplasm GIDX is found to represent the same germplasm as GIDY, or when GIDY contains correct or additional information compared to GIDX.
Checks:
a) Check for dependencies. If GIDX is a derivative and GIDY is from a
different germplasm group to GIDX then trace all germplasm in the same group as
GIDX with derivative paths which do not pass through GIDX as these will remain
unchanged in the change process and their relevance needs to be examined. b) Check that GIDX is later than GIDY, or all dependencies are later than GIDY. c) Check what names and attributes of GIDX should be copied to GIDY. d) If GIDX is generative and GIDY is derivative then GIDX must not occur
as a group source (GPID1 in derivative germplasm).
Changes:
a) There are four cases to distinguish:
Case 1. GIDX and GIDY both produced by generative processes
Case 2. GIDX and GIDY both derivatives in the SAME GERMPLASM GROUP
Case 3. GIDX is generative and GIDY is derivative in group GY
Case 4. GIDX is derivative and GIDY is generative or derivative but
from a different germplasm group to GIDX.
b) Obtain a date-time stamp for the change from the computer clock to be
added stored in the CGROUP field of change records for dependencies
as described below.
c) For cases 1 and 2, simply change all occurrences of GIDX in progenitor
fields (of the GERMPLASM table and the OTHER-PROGENITORS table) to GIDY using 3.9.3
and 3.9.6. (These occurrences are called dependencies).
For case 3, change the group IDs (PROGENITOR_ID1) of all derivatives of
GIDX to GY using 3.9.5 and change all other dependencies of GIDX to GIDY
using 3.9.3 or 3.9.6.
For case 4, change the group ID of all derivatives in the same group as
GIDX which have derivative paths tracing through GIDX to the group of GIDY
(GIDY itself if it is generative) using 3.9.5. Change all other dependencies of GIDX to GIDY
using 3.9.3 or 3.9.6.
d) Request reason for change.
e) Add a CHANGE record the CHANGES table with the following values:
CID - Next available negative integer
CTABLE - GERMPLSM
CFIELD - GID
CRECORD - GIDX
CFROM - GIDX
CTO - GIDY
CDATE - Current date
CTIME - Current time
CGROUP - date/time stamp for all dependant changes YYYYMMDDHHMMSSMM
CUID - Current user ID.
CSTATUS - 0
CDESC- Reason for change
f) If GIDX<0 set the GERMPLASM_REPLACE field of record GIDX to value GIDY.
Delete a Germplasm Record
This change may occur when it is discovered that a germplasm entry with GIDX is definitely wrong, but no alternative is known. If an alternative is known then this correction should not be used because it destroys all dependencies to the deleted germplasm. If the known alternative is already in the GMS then the replace GERMPLASM procedure should be used (3.9.8), otherwise fields of the current germplasm record should be changed to match the correct values (3.9.2-3.9.7).
Checks:
a) Check if GIDX has any dependencies – offspring, group subjects or
derivatives. (Check for GIDX in the PROGENITOR_ID1 and PROGENITOR_ID2 fields of the
GERMPLASM table and in the PROGENITOR_ID field of the OTHER_PROGENITORS table).
Changes:
a) Request a reason for change and obtain a date-time stamp for the deletion. b) Change GIDX to MISSING (0) in all dependencies using 3.9.4 – 3.9.6,
adding the same date-time stamp to the CGROUP field of the change record
for each dependency. c) Add a CHANGE record for GIDX exactly as in 3.9.8e except that CTO=GIDX. d) If GIDX<0, set the GERMPLASM_REPLACE field of GIDX to value GIDX.