GRIMS ICIS
From ICISWiki
GRIMS integration with ICIS
ICIS executable modules called internally from GRIMS
SetGen
GMSSrch
InTrack
ICIS DLL's called internally
GRIMS uses the dynamic link library ICIS32.DLL, which is written in C/ C++. The calls are made through a Delphi library (DelphiLib), which is shared among other ICIS applications written in Delphi. Below are the ICIS32 functions used by GRIMS:
| ICIS32 function | GRIMS module(s) |
|---|---|
| GMS_CloseDataBase | System |
| GMS_OpenDataBase2 | System |
| GMS_AddChanges | Acquisition, Seed Management |
| GMS_GetName | Acquisition, Seed Management |
| GMS_GetUser | Acquisition, Seed Management |
| GMS_AddName | Acquisition |
| GMS_SetName | Acquisition |
| GMS_ExpandDate | Acquisition, Seed Management, Characterization, Multiplication |
| GMS_ValidDate | System, Acquisition, Seed Management, Characterization, Multiplication |
| GMS_StrToDate | System, Acquisition, Seed Management, Characterization, Multiplication |
| GMS_StandardName | Acquisition |
| GMS_Commit | System, Acquisition, Seed Management, Characterization, Multiplication |
| GMS_Rollback | System, Acquisition, Seed Management, Characterization, Multiplication |
| GMS_GetListName | Acquisition, Seed Management, Characterization, Multiplication |
| GMS_AddListName | Acquisition, Seed Management, Characterization, Multiplication |
| GMS_SetListName | Acquisition, Seed Management, Characterization, Multiplication |
| DMS_openDatabase | System, Seed Management, Characterization, Multiplication |
| DMS_closeDatabase | System, Seed Management, Characterization, Multiplication |
| DMS_getScaleDis | Acquisition |
| DMS_getScaleDis2 | Acquisition |
| DMS_minOunit | Acquisition |
GRIMS specific query functions
Functions that query GRIMS specific tables are not available through the DelphiLib/ICIS32.dll libraries described above. A collection of such Delphi functions is shared by all GRIMS modules and maintained as a unit called IRGCISLibs.
ICIS modules called by GRIMS
- SetGen
- InTrack
ICIS sub-systems used by GRIMS
- lists
- user-defined fields
ICIS tables used by GRIMS
| ICIS Tables | GRIMS module(s) |
|---|---|
| GERMPLSM | Acquisition, Seed Management |
| LISTNMS | Acquisition, Seed Management, Characterization, Multiplication |
| LISTDATA | Acquisition, Seed Management, Characterization, Multiplication |
| NAMES | Acquisition, Seed Management, Characterization, Multiplication |
| ATTRIBUTE | Acquisition |
| UDFLDS | System, Acquisition, Seed Management, Characterization, Multiplication |
| LOCATION | System, Acquisition, Seed Management, Characterization, Multiplication |
| PERSONS | System, Acquisition, Seed Management, Characterization, Multiplication |
| IMS_LOT | System, Acquisition, Seed Management, Characterization, Multiplication |
| IMS_TRANSACTION | System, Acquisition, Seed Management, Characterization, Multiplication |
| TRAIT | Acquisition, Seed Management, Characterization, Multiplication |
| SCALE | Acquisition, Seed Management, Characterization, Multiplication |
| METHOD | Acquisition, Seed Management, Characterization, Multiplication |
| SCALETAB | Acquisition, Seed Management, Characterization, Multiplication |
| SCALECON | Acquisition, Seed Management, Characterization, Multiplication |
| SCALEDIS | Acquisition, Seed Management, Characterization, Multiplication |
Parallel Implementation
While the GRIMS application (based on Borland Delphi 6.0) is under development and only a part of its module has been released, Oracle developer based IRGCIS is still being used by the Genebank staff. For example, inventory adjustment is being done in GRIMS while the selection of planting material-which uses inventory data, runs in Oracle IRGCIS. A number of PL/SQL scripts are used to facilitate successful parallel implementation of the two systems.
Stored Procedures/ Functions/ SQL Scripts
Below are the stored procedures/functions/sql scripts that facilitate data transfer and/or automatic updates of GRC's IRIS local database.
Update IMS_LOT
Update NAMES
Initial Insert into NAMES
Initial Insert into ATRIBUTS
Storing Passport Data
Passport ERD using GMS and DMS Schema
Using the ATRIBUTS table for Passport data
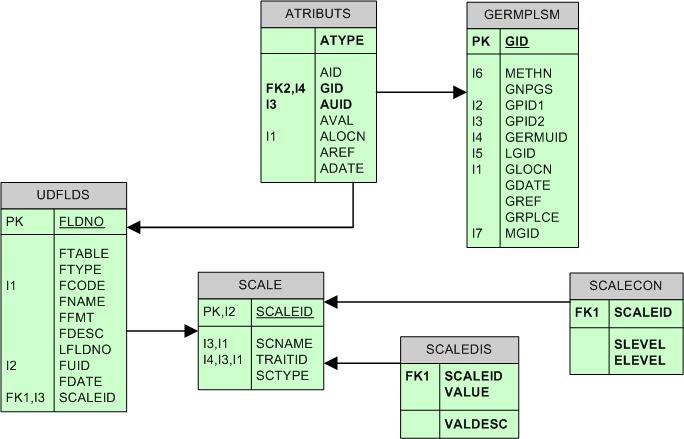
The ATTRIBUTES TABLE (ATRIBUTS)
ATRIBUTS table is used to store information about germplasm genesis, genealogy and nomenclature, which is not stored in other tables. This table also stores all the passport data of a germplasm.
| Columns - Short Name (Name) | Description | Type | Size (bytes) |
|---|---|---|---|
| ATTRIBUTE_ID (AID) | A unique sequential identifier | Number (Long) | 4 |
| GERMPLASM _ID (GID) | ID of germplasm with which the passport data is associated. GID in GERMPLASM table. | Number (Long) | 4 |
| ATTRIBUTE_TYPE (ATYPE) | Number that identifies the passport descriptor. FLDNO in UDFLDS table. Use 999 to mark the attribute as deleted. | Number (Integer). | 2 |
| ATTRIBUTE_USER_ID (AUID) | ID of the user giving the attribute value. UID in USERS table | Number (Integer) | 2 |
| ATTRIBUTE _VALUE (AVAL) | Contains the passport data | Text | 255 |
| ATTRIBUTE_LOCATION_NO (ALOCN) | Identifier for the location where the attribute value was set. LOCN on the LOCATION table. ALOCN = 0 if the location is unknown. TT Chang Genetic Resources Center has ALOCN=9016. | Number (Long) | 4 |
| ATTRIBUTE_REFERENCE(AREF) | Key to the reference for the attribute data. MISSING if the source is unknown. REFID in BIBREFS table. | Number (Long) | 4 |
| ATTRIBUTE_DATE (ADATE) | Date of value assignation. (YYYYMMDD) | Number (Long) | 4 |
Passport descriptors in UDFLDS table
Passport descriptors are defined in the UDFLDS table. The SCALEID field is linked to the SCALE table which determines the scale type of the passport descriptor (i.e. discrete, numeric, text, etc.). If the SCTYPE = "D" (meaning discrete) the valid values can be found in SCALEDIS. If SCTYPE="C" (continuous), the valid range can be found in SCALECON table.
Some of the attribute types already evolved in IRIS. For example, the FLDNO=1131 is originally assigned to store the FAO_DATE. FAO date is used by genebank before the year 2004 to determine whether the germplasm is FAO designated or not. It determines where or not an accession can be distributed or not. Today, the ATTRIBUTE name FAO_DATE is changed to IP_STAT to represent the IP status of a germplasm. In that way, other IRRI OU's can also use the attribute.
| FLDNO | FTABLE | FTYPE | FCODE | FNAME | FDESC | LFLDNO | FUID | FDATE | SCALEID |
|---|---|---|---|---|---|---|---|---|---|
| 1147 | ATRIBUTS | PASSPORT | BREEDSYS | Breeding system | Method of reproduction | 0 | 0 | 20041116 | -57 |
| 1136 | ATRIBUTS | PASSPORT | COLL_DATE | Date when the sample was collected | Date sample was collected
Format : DD-MON-YYYY
where : DD - day sample was collected
MON - month sample was collected
YYYY - year sample was collected
| 0 | 0 | 20041116 | -11 |
| 1135 | ATRIBUTS | PASSPORT | COLL_SOURCE | Collection source | Specific place where sample was collected | 0 | 0 | 20041116 | 527 |
| -9 | ATRIBUTS | PASSPORT | COLL_SOURCE_OTH | Collection source(specific) | 0 | 0 | 20041116 | -14 | |
| -2069 | ATRIBUTS | PASSPORT | COVER | Cover of wild rice (%) | % coverage (area) of wild rice at collecting site, estimated to the nearest 5% | 0 | 0 | 20041116 | -58 |
| 1539 | ATRIBUTS | PASSPORT | CULT_TYPE | Cultural type | Rice ecosystem | 0 | 0 | 20041116 | -16 |
| -2070 | ATRIBUTS | PASSPORT | DEG_GRAZE | Degree of grazing | A disturbance factor which refers to the extent of grazed area observed at the collection site | 0 | 0 | 20041116 | -59 |
| -2071 | ATRIBUTS | PASSPORT | DEG_INTROG | Degree of introgression | Spread of genes of one species into the gene complex of another | 0 | 0 | 20041116 | -60 |
| 1538 | ATRIBUTS | PASSPORT | DIRECT_SEED | Direct seeding | 0 | 0 | 20041116 | -17 | |
| 1510 | ATRIBUTS | PASSPORT | DIST_TO_SAT | Distance to o. sativa field | Distance of collection site of wild species to the nearest O. sativa field | 0 | 0 | 20041116 | -61 |
| 1537 | ATRIBUTS | PASSPORT | DOUBLE_TRANS | Double transplanting | 0 | 0 | 20041116 | -19 | |
| 1536 | ATRIBUTS | PASSPORT | DRAINAGE | Drainage | Manner in which the waters pass off the surface of the land | 0 | 0 | 20041116 | -20 |
| 1535 | ATRIBUTS | PASSPORT | ECOSYS | Ecological system | Refers to the environment where the sample was collected | 0 | 0 | 20041116 | -21 |
| 1534 | ATRIBUTS | PASSPORT | ECOZONE | Ecological zone | Refers to the edaphic-climatic condition of the area | 0 | 0 | 20041116 | -22 |
| 1131 | ATRIBUTS | ATTRIBUTE | FAO_DATE | Fao designation date | 0 | 0 | 20041116 | ||
| 1402 | ATRIBUTS | ATTRIBUTE | FAO_REMARKS | Fao remarks | 0 | 0 | 20041116 | ||
| -2073 | ATRIBUTS | PASSPORT | FLOATING | Floating | 0 | 0 | 20041116 | -62 | |
| 1509 | ATRIBUTS | PASSPORT | FLOWER | Flowering compared to o. sativa | Time of flowering of collected wild species sample compared to O. sativa | 0 | 0 | 20041116 | -63 |
| 1533 | ATRIBUTS | PASSPORT | FREQ | Frequency | Frequency of occurrence of species/variety collected in an area | 0 | 0 | 20041116 | -23 |
| 1532 | ATRIBUTS | PASSPORT | FUND | Funding agency | Source of funding | 0 | 0 | 20041116 | -24 |
| 1149 | ATRIBUTS | PASSPORT | GROWER | Grower's name | Name of farmer/grower of the sample collected | 0 | 0 | 20041116 | -25 |
| 1508 | ATRIBUTS | PASSPORT | GROWTH_STAGE | Growth stage | Growth stage of plant sampled during collection time | 0 | 0 | 20041116 | -64 |
| 1531 | ATRIBUTS | PASSPORT | HARVEST_DATE | Harvest date | Date variety was harvested | 0 | 0 | 20041116 | -26 |
| 1530 | ATRIBUTS | PASSPORT | HERB | Herbarium sample presence (y/n) | Indicates whether an herbarium sample was taken or not | 0 | 0 | 20041116 | -27 |
| 1529 | ATRIBUTS | PASSPORT | LANG_VAR | Language of variety name | Language/dialect of the varietal name or vernacular name of wild species | 0 | 0 | 20041116 | -28 |
| -2106 | ATRIBUTS | PASSPORT | LPCO_REV | Lemma and palea color | 0 | 0 | 20041116 | -66 | |
| 1529 | ATRIBUTS | PASSPORT | LANG_VAR | Language of variety name | Language/dialect of the varietal name or vernacular name of wild species | 0 | 0 | 20041116 | -28 |
| -2106 | ATRIBUTS | PASSPORT | LPCO_REV | Lemma and palea color | 0 | 0 | 20041116 | -66 | |
| 1528 | ATRIBUTS | PASSPORT | MAT | Maturity | 0 | 0 | 20041116 | -100 | |
| 1134 | ATRIBUTS | ATTRIBUTE | MISSION_CODE | Mission code | 0 | 0 | 20041116 | -77 | |
| 1527 | ATRIBUTS | PASSPORT | MIXED_STAND | Mixed_stand | 0 | 0 | 20041116 | -29 | |
| 223 | ATRIBUTS | ATTRIBUTE | MLS_DATE | Mls designation date | 0 | 0 | 20041116 | ||
| 1526 | ATRIBUTS | PASSPORT | MNG_VAR | Meaning of variety name in english | Translation of the vernacular name into English | 0 | 0 | 20041116 | -30 |
| 224 | ATRIBUTS | PASSPORT | ORI_COUNTRY | Country of origin | Country from which the sample originally came | 0 | 0 | 20041116 | -78 |
| -2107 | ATRIBUTS | PASSPORT | PHOTO_SENS | Photo sensitivity | 0 | 0 | 20041116 | -67 | |
| -2163 | ATRIBUTS | PASSPORT | PLT | Panicle length | 0 | 0 | 20041116 | -101 | |
| -2113 | ATRIBUTS | PASSPORT | POP_COMP | Population composition | The degree of homogeneity or heterogeneity of the sample | 0 | 0 | 20041116 | -68 |
| -2114 | ATRIBUTS | PASSPORT | POP_FERT | Population fertility | General seed set in a population per unit area | 0 | 0 | 20041116 | -69 |
| 1507 | ATRIBUTS | PASSPORT | POP_SIZE | Population size (sq. meters) | Population size in square meters, estimated visually at collection site | 0 | 0 | 20041116 | -70 |
| -2116 | ATRIBUTS | PASSPORT | PTY | Panicle type | 0 | 0 | 20041116 | 204 | |
| -2117 | ATRIBUTS | PASSPORT | RATOON | Ratoon | Presence of tillers from nodes | 0 | 0 | 20041116 | -71 |
| 1525 | ATRIBUTS | PASSPORT | REM_GRAIN | Grain characteristics | Grain characteristics as observed by the collector | 0 | 0 | 20041116 | -32 |
| 1524 | ATRIBUTS | ATTRIBUTE | REM_LAT_LONGI | Remark on lat/long (location) | 0 | 0 | 20041116 | ||
| 1403 | ATRIBUTS | PASSPORT | REM_OTHER | Other observations of the collector | Other observations of the collector | 0 | 0 | 20041116 | -32 |
| -40 | ATRIBUTS | PASSPORT | REM_PEST | Reaction to pests/diseases | Resistance to pests/diseases | 0 | 0 | 20041116 | -32 |
| 1523 | ATRIBUTS | PASSPORT | REM_PLANT | Plant characteristics | Plant characteristics as observed by the collector | 0 | 0 | 20041116 | -32 |
| 1400 | ATRIBUTS | ATTRIBUTE | REM_SAMPLE_STAT | Remark on sample status | 0 | 0 | 20041116 | ||
| 1148 | ATRIBUTS | PASSPORT | REM_SPECIAL | Special characteristics | Special characteristics of the sample collected | 0 | 0 | 20041116 | -32 |
| 1144 | ATRIBUTS | PASSPORT | SAMPLE_STAT | Status of sample | Type of germplasm collected | 0 | 0 | 20041116 | -36 |
| 1139 | ATRIBUTS | PASSPORT | SAMPLE_STAT_OTH | Sample status (specific) | 0 | 0 | 20041116 | -37 | |
| 1140 | ATRIBUTS | PASSPORT | SAMPLE_TYPE | Type of sample | Plant parts collected | 0 | 0 | 20041116 | -38 |
| 1520 | ATRIBUTS | PASSPORT | SAMP_METHOD | Sampling method | Indicates how the collected material were sampled | 0 | 0 | 20041116 | -33 |
| 1521 | ATRIBUTS | PASSPORT | SAMP_METHOD_SP | Sampling method | 0 | 0 | 20041116 | -34 | |
| 1146 | ATRIBUTS | PASSPORT | SAMP_ORIG | Sample origin | Indicates whether a sample is indigenous or introduced to the area where it was collected | 0 | 0 | 20041116 | -35 |
| -2118 | ATRIBUTS | PASSPORT | SCCO_REV | Seed coat color | 0 | 0 | 20041116 | -72 | |
| 1519 | ATRIBUTS | PASSPORT | SEED_FILE | Seed file presence | 0 | 0 | 20041116 | -79 | |
| 1505 | ATRIBUTS | PASSPORT | SEED_PROD | Seed production | Seed production | 0 | 0 | 20041116 | -73 |
| 1142 | ATRIBUTS | ATTRIBUTE | SENDER | Sender's name | Name of sender (can be deleted) | 0 | 0 | 20041116 | |
| 1504 | ATRIBUTS | PASSPORT | SHADE | Shading | Refers to the amount of exposure to the sun of the plant where sample was collected | 0 | 0 | 20041116 | -74 |
| 1516 | ATRIBUTS | PASSPORT | SHIFT_CULT | Shifting culture | 0 | 0 | 20041116 | -39 | |
| 1518 | ATRIBUTS | PASSPORT | SITE | Site | Topographical condition of a specific area where sample was collected | 0 | 0 | 20041116 | -40 |
| 1517 | ATRIBUTS | PASSPORT | SOIL_TEXT | Soil texture | Relative proportions of sand, silt and clay in a sample | 0 | 0 | 20041116 | -44 |
| 1501 | ATRIBUTS | PASSPORT | SOURCE_INFO | Source of information | Refers to the person or publication where the passport information was obtained | 0 | 0 | 20041116 | -41 |
| 1515 | ATRIBUTS | PASSPORT | SOW_DATE | Sowing date | Date sample was sown | 0 | 0 | 20041116 | -42 |
| 1133 | ATRIBUTS | PASSPORT | SPP_CODE | Species code | 0 | 0 | 20041116 | -84 | |
| 1503 | ATRIBUTS | PASSPORT | SPP_DIVER | Species diversity | Approximate number of species in the collection site | 0 | 0 | 20041116 | -75 |
| 1540 | ATRIBUTS | ATTRIBUTE | STATUS_ACC | Status_acc | 0 | 0 | 20041116 | -1013 | |
| 1141 | ATRIBUTS | PASSPORT | TAXNO | Taxonomy | 0 | 0 | 20041116 | -87 | |
| 1514 | ATRIBUTS | PASSPORT | TERRACED_CULT | Terraced culture | 0 | 0 | 20041116 | -45 | |
| 1513 | ATRIBUTS | PASSPORT | TOPO | Topography | Configuration of a surface including its relief and the position of its natural and man-made features | 0 | 0 | 20041116 | -46 |
| 1512 | ATRIBUTS | PASSPORT | TOPO_OTH | Topography (others) | 0 | 0 | 20041116 | -47 | |
| 1511 | ATRIBUTS | PASSPORT | TRANS_DATE | Transplanting date | Date sample was transplanted | 0 | 0 | 20041116 | -49 |
| 1143 | ATRIBUTS | PASSPORT | USAGE | Usage of variety collected | Usage of variety collected | 0 | 0 | 20041116 | -50 |
| 1145 | ATRIBUTS | PASSPORT | VAR_SAMP | Varietal sample | Describes the composition of the variety collected | 0 | 0 | 20041116 | -51 |
| 1132 | ATRIBUTS | PASSPORT | VG | Varietal group | 0 | 0 | 20041116 | -52 | |
| 1502 | ATRIBUTS | PASSPORT | WATER_DEPTH | Water depth (m) | Approximate depth of water where sample was collected | 0 | 0 | 20041116 | -76 |
* The germplasm taxonomy and mission information are stored in a separate table.
Negative scales are still pending for uploading to central. Request was submitted in April 2007. http://cropforge.org/tracker/index.php?group_id=82&atid=418.
--rherrera 10:28, 4 August 2008 (PHT)
Discrete values of passport data (SCALEDIS)
| Attribute type (FLDNO) | Passport Descriptor (FCODE) | SCALEID | Valid Value (VALUE) | Description (VALDESC) |
|---|---|---|---|---|
| -2067 | AWN_STRENGTH | -56 | 1 | SOFT |
| -2067 | AWN_STRENGTH | -56 | 2 | HARD |
| -2067 | AWN_STRENGTH | -56 | 3 | INTERMEDIATE |
| -2068 | BREEDSYS | -57 | 1 | INBREEDING |
| -2068 | BREEDSYS | -57 | 999 | MIXTURE |
| -2068 | BREEDSYS | -57 | 4 | UNSURE |
| -2068 | BREEDSYS | -57 | 3 | VEGETATIVE |
| -2068 | BREEDSYS | -57 | 2 | OUTCROSSING |
| -8 | COLL_SOURCE | -13 | 1 | FARMLAND |
| -8 | COLL_SOURCE | -13 | 9 | OTHER |
| -8 | COLL_SOURCE | -13 | 4 | VILLAGE MARKET |
| -8 | COLL_SOURCE | -13 | 5 | COMMERCIAL MARKET |
| -8 | COLL_SOURCE | -13 | 6 | INSTITUTE |
| -8 | COLL_SOURCE | -13 | 8 | WILD |
| -8 | COLL_SOURCE | -13 | 7 | FIELD BORDER |
| -8 | COLL_SOURCE | -13 | 3 | FARMSTORE |
| -8 | COLL_SOURCE | -13 | 2 | THRESHING FLOOR |
| -11 | CULT_TYPE | -16 | 1 | IRRIGATED |
| -11 | CULT_TYPE | -16 | 4 | UPLAND |
| -11 | CULT_TYPE | -16 | 5 | TIDAL WETLAND |
| -11 | CULT_TYPE | -16 | 6 | SWAMP |
| -11 | CULT_TYPE | -16 | 2 | RAINFED-LOWLAND |
| -11 | CULT_TYPE | -16 | 3 | DEEPWATER |
| -2070 | DEG_GRAZE | -59 | 1 | NO GRAZING |
| -2070 | DEG_GRAZE | -59 | 3 | 50% GRAZED |
| -2070 | DEG_GRAZE | -59 | 5 | ALMOST COMPLETE GRAZING (ABOVE 80%) |
| -2070 | DEG_GRAZE | -59 | 4 | 75% GRAZED |
| -2070 | DEG_GRAZE | -59 | 2 | 25% GRAZED |
| -2071 | DEG_INTROG | -60 | 1 | NO INTROGRESSION |
| -2071 | DEG_INTROG | -60 | 9 | ALMOST THE SAME AS O. SATIVA |
| -2071 | DEG_INTROG | -60 | 4 | 6-10% GENE FLOW FROM O. SATIVA TO WILD POP. |
| -2071 | DEG_INTROG | -60 | 5 | MANY PLANTS IN POP. ARE LIKE O. SATIVA |
| -2071 | DEG_INTROG | -60 | 6 | 20-40% GENE FLOW FROM O. SATIVA TO WILD POP. |
| -2071 | DEG_INTROG | -60 | 8 | 60-80% GENE FLOW FROM O. SATIVA TO WILD POP. |
| -2071 | DEG_INTROG | -60 | 7 | HEAVILY INTROGRESSED FROM O. SATIVA |
| -2071 | DEG_INTROG | -60 | 3 | FEW PLANTS IN POP. WITH O. SATIVA CHARACTERS (2-5%) |
| -2071 | DEG_INTROG | -60 | 2 | < 1% GENE FLOW FROM O. SATIVA TO WILD POP. |
| -12 | DIRECT_SEED | -17 | N | NO |
| -12 | DIRECT_SEED | -17 | Y | YES |
| -2072 | DIST_TO_SAT | -61 | 1 | 0 (IN CULTIVATED FIELD) |
| -2072 | DIST_TO_SAT | -61 | 4 | FAR(NOT EASILY ACCESSIBLE FR.COLLECTING SITE) |
| -2072 | DIST_TO_SAT | -61 | 2 | <= 20 M |
| -2072 | DIST_TO_SAT | -61 | 3 | > 20 M |
| -14 | DOUBLE_TRANS | -19 | N | NO |
| -14 | DOUBLE_TRANS | -19 | Y | YES |
| -15 | DRAINAGE | -20 | 1 | POOR |
| -15 | DRAINAGE | -20 | 3 | GOOD |
| -15 | DRAINAGE | -20 | 4 | EXCESSIVE |
| -15 | DRAINAGE | -20 | 2 | MODERATE |
| -16 | ECOSYS | -21 | 1 | DRYLAND |
| -16 | ECOSYS | -21 | 3 | SHALLOW FLOODED SWAMP |
| -16 | ECOSYS | -21 | 5 | FLOATING RICE |
| -16 | ECOSYS | -21 | 7 | IRRIGATION |
| -16 | ECOSYS | -21 | 6 | MANGROVE |
| -16 | ECOSYS | -21 | 4 | DEEP FLOODED SWAMP |
| -16 | ECOSYS | -21 | 2 | HYDROMORPHIC |
| -17 | ECOZONE | -22 | 1 | FOREST |
| -17 | ECOZONE | -22 | 2 | TRANSITION ZONE |
| -17 | ECOZONE | -22 | 3 | DERIVED SAVANNA |
| -17 | ECOZONE | -22 | 5 | SUDAN SAVANNA |
| -17 | ECOZONE | -22 | 7 | MONTANE/HIGHLAND |
| -17 | ECOZONE | -22 | 6 | SAHEL (SEMI-DESERT) |
| -17 | ECOZONE | -22 | 4 | GUINEA SAVANNA |
| -2073 | FLOATING | -62 | 3 | INTERMEDIATE |
| -2073 | FLOATING | -62 | N | NO |
| -2073 | FLOATING | -62 | Y | YES |
| -2074 | FLOWER | -63 | 1 | EARLIER THAN O. SATIVA |
| -2074 | FLOWER | -63 | 5 | LATER THAN O. SATIVA |
| -2074 | FLOWER | -63 | 2 | RANGES FROM BEFORE O. SAT. TO SAME AS O. SAT. |
| -2074 | FLOWER | -63 | 4 | RANGES FROM SAME AS O. SAT. TO AFTER O. SAT. |
| -2074 | FLOWER | -63 | 3 | SAME AS O. SATIVA |
| -22 | FREQ | -23 | 1 | ABUNDANT |
| -22 | FREQ | -23 | 2 | FREQUENT |
| -22 | FREQ | -23 | 3 | OCCASIONAL |
| -22 | FREQ | -23 | 4 | RARE |
| -2085 | GROWTH_STAGE | -64 | 1 | VEGETATIVE |
| -2085 | GROWTH_STAGE | -64 | 4 | SEED SHED |
| -2085 | GROWTH_STAGE | -64 | 2 | FLOWERING |
| -2085 | GROWTH_STAGE | -64 | 3 | MATURE |
| -26 | HERB | -27 | N | NO |
| -26 | HERB | -27 | Y | YES |
| -2145 | LAT_DIR | -90 | N | NORTH |
| -2145 | LAT_DIR | -90 | S | SOUTH |
| -2149 | LONGI_DIR | -94 | E | EAST |
| -2149 | LONGI_DIR | -94 | W | WEST |
| -2106 | LPCO_REV | -66 | 010 | WHITE |
| -2106 | LPCO_REV | -66 | 020 | STRAW |
| -2106 | LPCO_REV | -66 | 042 | GOLD AND GOLD FURROWS |
| -2106 | LPCO_REV | -66 | 052 | BROWN (TAWNY) |
| -2106 | LPCO_REV | -66 | 053 | BROWN SPOTS |
| -2106 | LPCO_REV | -66 | 054 | BROWN FURROWS |
| -2106 | LPCO_REV | -66 | 080 | PURPLE |
| -2106 | LPCO_REV | -66 | 082 | REDDISH TO LIGHT PURPLE |
| -2106 | LPCO_REV | -66 | 090 | PURPLE SPOTS |
| -2106 | LPCO_REV | -66 | 091 | PURPLE FURROWS |
| -2106 | LPCO_REV | -66 | 100 | BLACK |
| -2106 | LPCO_REV | -66 | 999 | MIXTURE |
| -2145 | LAT_DIR | -90 | N | NORTH |
| -2145 | LAT_DIR | -90 | S | SOUTH |
| -2149 | LONGI_DIR | -94 | E | EAST |
| -2149 | LONGI_DIR | -94 | W | WEST |
| -2106 | LPCO_REV | -66 | 010 | WHITE |
| -2106 | LPCO_REV | -66 | 020 | STRAW |
| -2106 | LPCO_REV | -66 | 042 | GOLD AND GOLD FURROWS |
| -2106 | LPCO_REV | -66 | 052 | BROWN (TAWNY) |
| -2106 | LPCO_REV | -66 | 053 | BROWN SPOTS |
| -2106 | LPCO_REV | -66 | 054 | BROWN FURROWS |
| -2106 | LPCO_REV | -66 | 080 | PURPLE |
| -2106 | LPCO_REV | -66 | 082 | REDDISH TO LIGHT PURPLE |
| -2106 | LPCO_REV | -66 | 090 | PURPLE SPOTS |
| -2106 | LPCO_REV | -66 | 091 | PURPLE FURROWS |
| -2106 | LPCO_REV | -66 | 100 | BLACK |
| -2106 | LPCO_REV | -66 | 999 | MIXTURE |
| -30 | MIXED_STAND | -29 | N | NO |
| -30 | MIXED_STAND | -29 | Y | YES |
| -2107 | PHOTO_SENS | -67 | 3 | UNSURE |
| -2107 | PHOTO_SENS | -67 | N | NO |
| -2107 | PHOTO_SENS | -67 | Y | YES |
| -2113 | POP_COMP | -68 | 1 | HETEROGENEOUS |
| -2113 | POP_COMP | -68 | 2 | HOMOGENEOUS |
| -2114 | POP_FERT | -69 | 0 | NO SEEDS AVAILABLE |
| -2114 | POP_FERT | -69 | 1 | ABOUT 1% SEEDS AVAILABLE FROM MOST PLANTS |
| -2114 | POP_FERT | -69 | 2 | 2-5% SEEDS AVAILABLE FROM MOST PLANTS |
| -2114 | POP_FERT | -69 | 3 | FEW SEEDS AVAILABLE FROM MOST PLANTS (5-10%) |
| -2114 | POP_FERT | -69 | 4 | 10-20% SEEDS AVAILABLE FROM MOST PLANTS |
| -2114 | POP_FERT | -69 | 5 | W/ STERILE SPIKELETS BUT MOST ARE FERTILE SEEDS (20-40%) |
| -2114 | POP_FERT | -69 | 6 | 40-60% SEEDS AVAILABLE FROM MOST PLANTS |
| -2114 | POP_FERT | -69 | 7 | FEW STERILE SPIKELETS (60-80% FERTILE) |
| -2114 | POP_FERT | -69 | 8 | 80-99% FERTILE SPIKELETS |
| -2114 | POP_FERT | -69 | 9 | 100% FERTILE SPIKELETS |
| -2116 | PTY | -216 | 1 | COMPACT (1) |
| -2116 | PTY | -216 | 2 | COMPACT (2) |
| -2116 | PTY | -216 | 3 | COMPACT (3) |
| -2116 | PTY | -216 | 4 | INTERMEDIATE (4) |
| -2116 | PTY | -216 | 5 | INTERMEDIATE (5) |
| -2116 | PTY | -216 | 6 | INTERMEDIATE (6) |
| -2116 | PTY | -216 | 7 | OPEN (7) |
| -2116 | PTY | -216 | 8 | OPEN (8) |
| -2116 | PTY | -216 | 9 | OPEN (9) |
| -2116 | PTY | -216 | 999 | MIXTURE |
| -2117 | RATOON | -71 | 1 | MANY |
| -2117 | RATOON | -71 | 2 | FEW |
| -2118 | SCCO_REV | -72 | 010 | WHITE |
| -2118 | SCCO_REV | -72 | 050 | BROWN |
| -2118 | SCCO_REV | -72 | 051 | LIGHT BROWN |
| -2118 | SCCO_REV | -72 | 055 | SPECKLED BROWN |
| -2118 | SCCO_REV | -72 | 070 | RED |
| -2118 | SCCO_REV | -72 | 080 | PURPLE |
| -2118 | SCCO_REV | -72 | 088 | VARIABLE PURPLE |
| -2118 | SCCO_REV | -72 | 999 | MIXTURE |
| -2119 | SEED_PROD | -73 | 1 | LOW |
| -2119 | SEED_PROD | -73 | 2 | HIGH |
| -2120 | SHADE | -74 | 1 | OPEN |
| -2120 | SHADE | -74 | 2 | PARTIAL SHADE |
| -2120 | SHADE | -74 | 3 | COMPLETE SHADE |
| -44 | SAMPLE_STAT | -36 | 1 | WILD |
| -44 | SAMPLE_STAT | -36 | 2 | WEEDY |
| -44 | SAMPLE_STAT | -36 | 3 | PRIMITIVE CULTIVAR |
| -44 | SAMPLE_STAT | -36 | 4 | OTHER |
| -46 | SAMPLE_TYPE | -38 | 1 | SEEDS |
| -46 | SAMPLE_TYPE | -38 | 2 | PANICLES |
| -46 | SAMPLE_TYPE | -38 | 3 | VEGETATIVE |
| -47 | SAMP_METHOD | -33 | 1 | RANDOM |
| -47 | SAMP_METHOD | -33 | 2 | NON-RANDOM |
| -49 | SAMP_ORIG | -35 | 1 | LOCAL(INDIGENOUS) |
| -49 | SAMP_ORIG | -35 | 2 | EXOTIC(INTRODUCED) |
| -50 | SEED_FILE | -79 | N | SEED FILE NOT AVAILABLE |
| -50 | SEED_FILE | -79 | Y | SEED FILE PRESENT |
| -56 | SITE | -40 | 1 | LEVEL |
| -56 | SITE | -40 | 2 | SLOPE |
| -56 | SITE | -40 | 3 | SUMMIT |
| -56 | SITE | -40 | 4 | DEPRESSION |
| -57 | SOIL_TEXT | -44 | 1 | SAND |
| -57 | SOIL_TEXT | -44 | 2 | LOAM |
| -57 | SOIL_TEXT | -44 | 3 | CLAY |
| -57 | SOIL_TEXT | -44 | 4 | SILT |
| -57 | SOIL_TEXT | -44 | 5 | HIGHLY ORGANIC |
| -58 | SHIFT_CULT | -39 | N | NO |
| -58 | SHIFT_CULT | -39 | Y | YES |
| -2121 | SPP_DIVER | -75 | 1 | SINGLE SPECIES (E.G. CULTIVATED FIELD) |
| -2121 | SPP_DIVER | -75 | 2 | IN AND AT THE EDGE OF A CULTIVATED FIELD AND A WEEDY HABITAT |
| -2121 | SPP_DIVER | -75 | 3 | FEW PREDOMINANT SPECIES (3-5) (WEEDY HABITAT) |
| -2121 | SPP_DIVER | -75 | 4 | WITHIN A WEEDY HABITAT AND A SEC. FOREST |
| -2121 | SPP_DIVER | -75 | 5 | BET. 5-10 PLANTS/SPP.(YOUNG SEC. FOREST) |
| -2121 | SPP_DIVER | -75 | 6 | SCATTERED WITHIN A YOUNG SEC. FOREST AND WELL-DEV. FOREST |
| -2121 | SPP_DIVER | -75 | 7 | MANY SPP.(10-20 SPP.) (WELL-DEV. SEC. FOREST) |
| -2121 | SPP_DIVER | -75 | 8 | ABUNDANT IN BOTH WELL-DEV. SEC. FOREST AND A CLIMAX COMMUNITY |
| -2121 | SPP_DIVER | -75 | 9 | ABUNDANT SPP.(>20 SPP.) (CLIMAX COMMUNITY) |
| -61 | SPP_CODE | -84 | G | O. GLABERRIMA |
| -61 | SPP_CODE | -84 | S | O. SATIVA |
| -61 | SPP_CODE | -84 | W | WILD SPECIES |
| -65 | STATUS_ACC | -1028 | AV | AVAILABLE |
| -65 | STATUS_ACC | -1028 | NA | NOT AVAILABLE |
| -67 | TERRACED_CULT | -45 | N | NO |
| -67 | TERRACED_CULT | -45 | Y | YES |
| -68 | TOPO | -46 | 1 | SWAMP |
| -68 | TOPO | -46 | 2 | FLOOD PLAIN |
| -68 | TOPO | -46 | 3 | PLAIN LEVEL |
| -68 | TOPO | -46 | 4 | UNDULATING |
| -68 | TOPO | -46 | 5 | HILLY |
| -68 | TOPO | -46 | 6 | MOUNTAINOUS |
| -68 | TOPO | -46 | 7 | OTHER |
| -73 | VARLINE_TYPE | -85 | I | IMPROVED VARIETY (ADV. CULTIVARS/ SELECTION) |
| -73 | VARLINE_TYPE | -85 | P | BREEDING AND INBRED LINES (PROMISING LINE) |
| -73 | VARLINE_TYPE | -85 | T | TRADITIONAL VARIETY (LANDRACES) |
| -75 | VAR_SAMP | -51 | 1 | SINGLE VARIETY |
| -75 | VAR_SAMP | -51 | 2 | VARIETAL MIXTURE |
| -76 | VG | -227 | 1 | INDICA |
| -76 | VG | -227 | 2 | JAPONICA |
| -76 | VG | -227 | 3 | JAVANICA |
| -76 | VG | -227 | 4 | INTERMEDIATE (HYBRIDS) |
| -76 | VG | -52 | 1 | INDICA |
| -76 | VG | -52 | 2 | JAPONICA |
| -76 | VG | -52 | 3 | JAVANICA |
| -76 | VG | -52 | 4 | INTERMEDIATE (HYBRIDS) |
Range of allowed values for numeric passport data (SCALECON)
| Attribute type (FLDNO) | Passport Descriptor (FCODE) | SCALEID | Minimum value | Maximum value |
|---|---|---|---|---|
| -2069 | COVER | -58 | 0 | 100 |
| -2114 | POP_FERT | -69 | 0 | 100 |
| -2115 | POP_SIZE | -70 | 0 | 80000 |
| -2122 | WATER_DEPTH | -76 | 0 | 5000 |
| -2144 | LAT_DEG | -89 | 0 | 90 |
| -2146 | LAT_MIN | -91 | 0 | 59 |
| -2148 | LONGI_DEG | -93 | 0 | 180 |
| -2150 | LONGI_MIN | -95 | 0 | 59 |
| -2151 | LONGI_SEC | -96 | 0 | 59 |
| -2153 | AWLT | -55 | 0 | 202 |
| -2163 | PLT | -101 | 3 | 50 |
| -28 | MAT | -100 | 0 | 300 |
User defined sql statements for storing passport data (SCALETAB)
| Attribute type (FLDNO) | Passport Descriptor (FCODE) | SCALEID | SQL Statement | Module where the tables can be found |
|---|---|---|---|---|
| -29 | MISSION_CODE | -77 | SELECT MISSION_CODE AS SAVEVAL,
ifnull(START_MISS,'-') START, ifnull(END_MISS,'-') END, INSTITUTID AS INSTITUT, ISO AS COUNTRY FROM IRGC_MISSION WHERE START_MISS IS NOT NULL ORDER BY START_MISS, END_MISS | DMSLOC |
| -34 | ORI_COUNTRY | -78 | SELECT LABBR AS SAVEVAL, LABBR AS 'FAO NAME',LNAME AS COUNTRY FROM LOCATION WHERE LTYPE=405 ORDER BY LNAME | DMSLOC |
| -52 | SENDER_CCODE | -4 | SELECT CONCAT(P.LNAME, ', ', P.FNAME) as SAVEVAL,
CONCAT(P.LNAME, ', ', P.FNAME) NAME, I.INSNAME AS INSTITUTE FROM PERSONS P INNER JOIN INSTITUT I ON P.INSTITID = I.INSTITID WHERE P.LNAME <> '-' ORDER BY LNAME | DMSLOC |
| -61 | SPP_CODE | -84 | SELECT SPECIES_COLL AS SAVEVAL, SPECIES_COLL AS 'SPECIES NAME', GENUS FROM SPECIES | DMSLOC |
| -62 | SS_COUNTRY | -6 | SELECT LABBR AS SAVEVAL, LABBR AS 'FAO NAME',
LNAME AS COUNTRY FROM LOCATION WHERE LTYPE=405 AND LNAME<> ORDER BY LNAME | DMSLOC |
GRIMS data uploaded to IRIS Central
GMS
- 117,384 accessions (i.e. including dead plant, not available for distribution accession) have a positive germplsm identifier.
- 117,284 names with name type =1 stored in the central IRIS
- 164,520 generations identified and assigned new (+) GIDs
- Passport data for 8 descriptors (The data were loaded between September 2004-July 2008.)
- Collection date- (COLL_DATE)
- Collection source- (COLL_SOURCE)
- Country of origin- (ORI_COUNTRY)
- Mls designation date- (MLS_DATE)
- FAO designation date- (FAO_DATE)
- Species code- (SPP_CODE)
- Taxonomy- (TAXNO)
- Varietal group- (VG)
Grims data for uploading to IRIS central
- Passport data for 115,632 accession for the following descriptors
- Cultivated & Wild species passport data
- Collection source(specific)- (COLL_SOURCE_OTH)
- Cultural type- (CULT_TYPE)
- Direct seeding- (DIRECT_SEED)
- District- (DISTRICT)
- Double transplanting- (DOUBLE_TRANS)
- Drainage- (DRAINAGE)
- Ecological system- (ECOSYS)
- Ecological zone- (ECOZONE)
- Fao designated seed- (FAO_PROC)
- Fao remarks- (FAO_REMARKS)
- Frequency- (FREQ)
- Funding agency- (FUND)
- Grower's name- (GROWER)
- Harvest date- (HARVEST_DATE)
- Herbarium sample presence (y/n)- (HERB)
- Language of variety name- (LANG_VAR)
- Maturity- (MAT)
- Mission code- (MISSION_CODE)
- Mixed_stand- (MIXED_STAND)
- Mls designated- (MLS_PROC)
- Meaning of variety name in english- (MNG_VAR)
- Previous designation- (PREV_NAME)
- Province- (PROV)
- Grain characteristics- (REM_GRAIN)
- Remark on lat/long (location)- (REM_LAT_LONGI)
- Other observations of the collector- (REM_OTHER)
- Reaction to pests/diseases- (REM_PEST)
- Plant characteristics- (REM_PLANT)
- Remark on sample status- (REM_SAMPLE_STAT)
- Special characteristics- (REM_SPECIAL)
- Status of sample- (SAMPLE_STAT)
- Sample status (specific)- (SAMPLE_STAT_OTH)
- Type of sample- (SAMPLE_TYPE)
- Sampling method- (SAMP_METHOD)
- Sampling method- (SAMP_METHOD_SP)
- Sample origin- (SAMP_ORIG)
- Shifting culture- (SHIFT_CULT)
- Site- (SITE)
- Soil texture- (SOIL_TEXT)
- Source of information- (SOURCE_INFO)
- Sowing date- (SOW_DATE)
- Source country- (SS_COUNTRY)
- Source station- (SS_STATION)
- Status_acc- (STATUS_ACC)
- Terraced culture- (TERRACED_CULT)
- Topography- (TOPO)
- Topography (others)- (TOPO_OTH)
- Town- (TOWN)
- Transplanting date- (TRANS_DATE)
- Usage of variety collected- (USAGE)
- Classification of o. sativa samples- (VARLINE_TYPE)
- Varietal sample- (VAR_SAMP)
- Village- (VILLAGE)
- Cultivated & Wild species passport data
- Wild species passport data
- Anther length-(ANLT)
- Awn length-(AWLT)
- Awn strength-(AWN_STRENGTH)
- Breeding system-(BREEDSYS)
- Cover of wild rice (%)-(COVER)
- Degree of grazing-(DEG_GRAZE)
- Degree of introgression-(DEG_INTROG)
- Distance to o. sativa field-(DIST_TO_SAT)
- Floating-(FLOATING)
- Flowering compared to o. sativa-(FLOWER)
- Grain length-(GRLT)
- Growth stage-(GROWTH_STAGE)
- Grain thickness-(GRTH)
- Grain width-(GRWD)
- Lemma and palea color-(LPCO_REV)
- Photo sensitivity-(PHOTO_SENS)
- Panicle length-(PLT)
- Population composition-(POP_COMP)
- Population fertility-(POP_FERT)
- Population size (sq. meters)-(POP_SIZE)
- Panicle type-(PTY)
- Ratoon-(RATOON)
- Seed coat color-(SCCO_REV)
- Seed production-(SEED_PROD)
- Shading-(SHADE)
- Species diversity-(SPP_DIVER)
- Water depth (m)-(WATER_DEPTH)
- Uploading of the metadata tables to the central DMS
- TRAIT
- SCALE
- METHODS
- SCALEDIS
- Uploading of the the characterization data to the central DMS